Development of User-Friendly Method to Distinguish Subspecies of the Korean Medicinal Herb Perilla frutescens Using Multiplex-PCR
Abstract
:1. Introduction
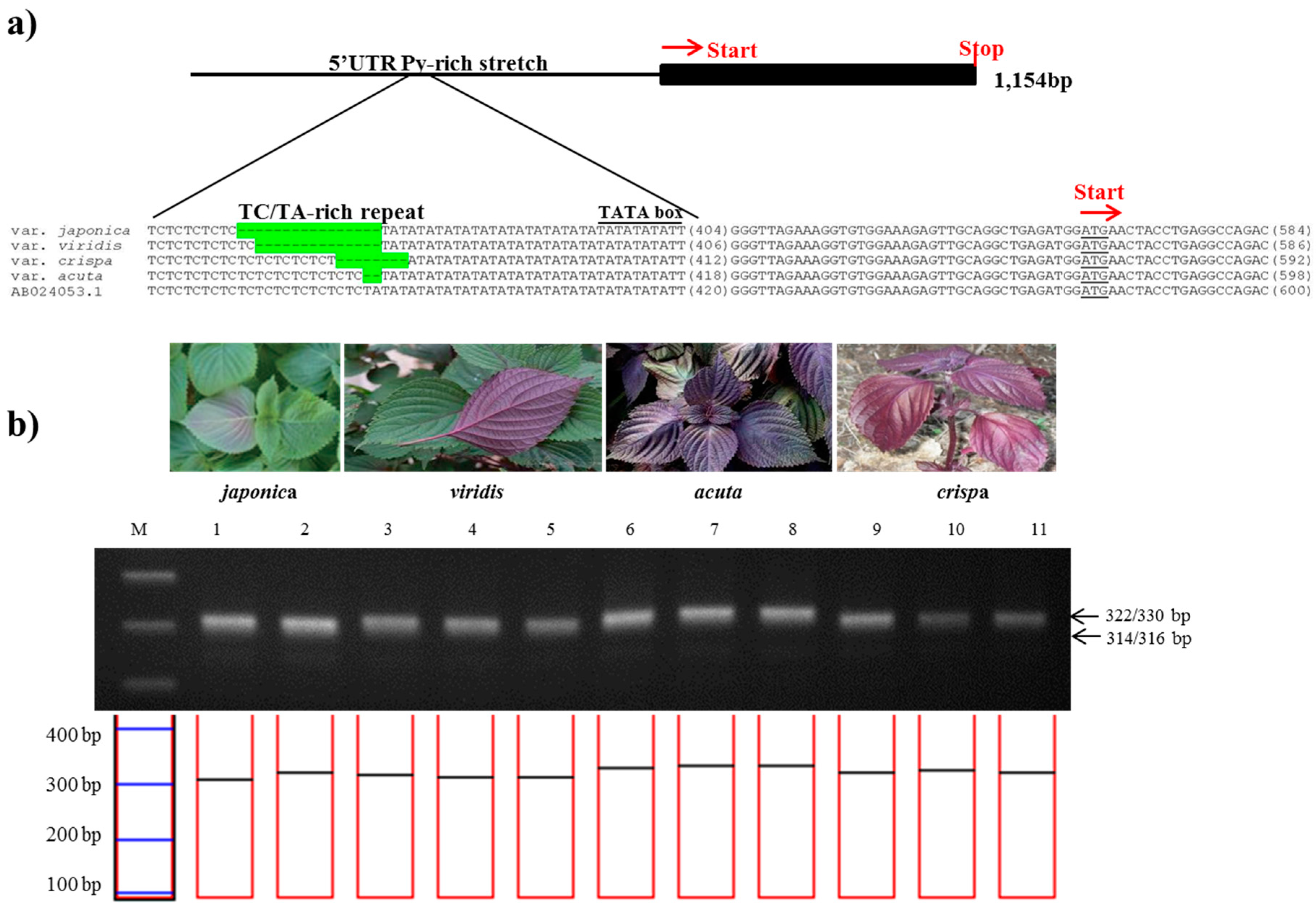
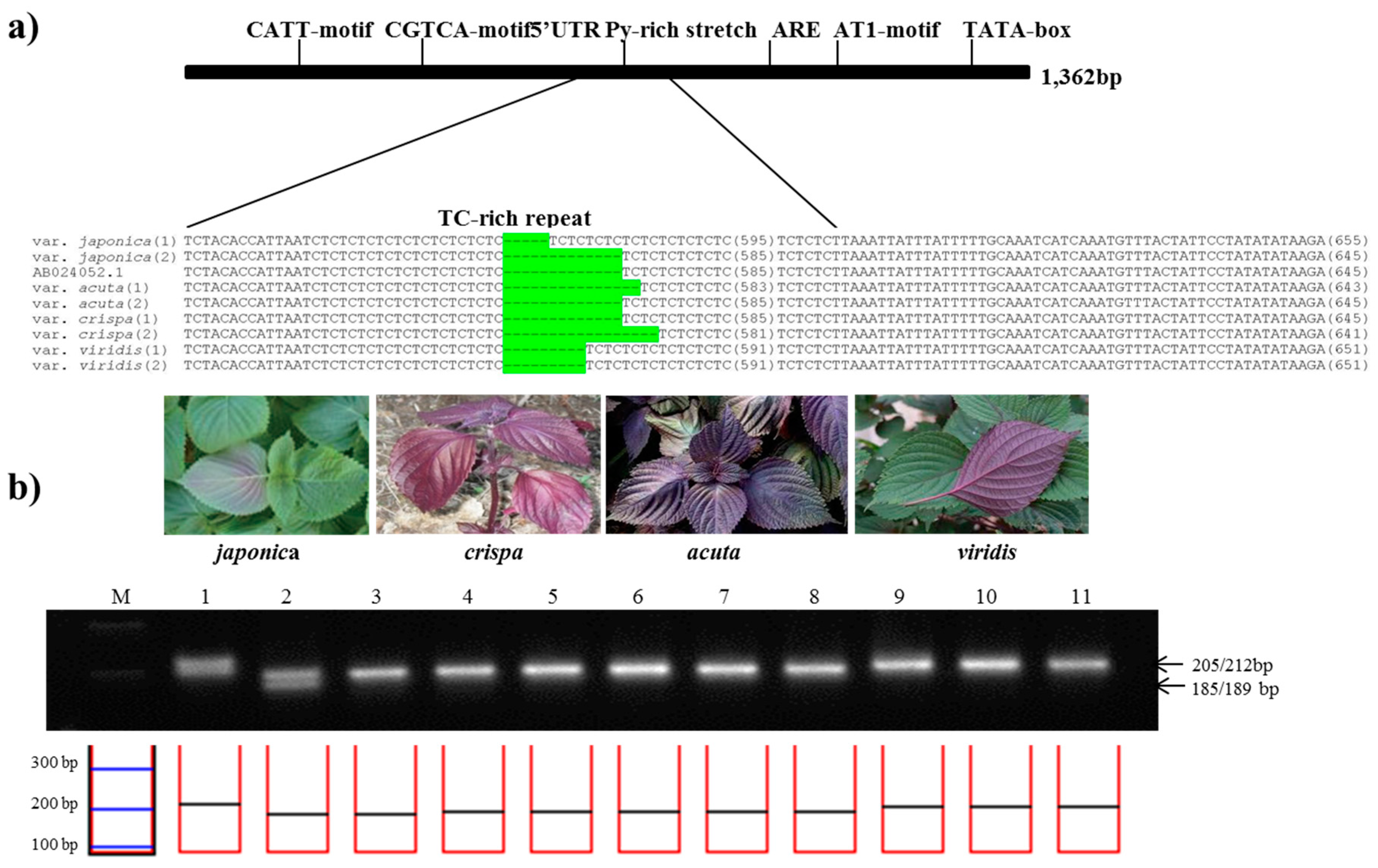
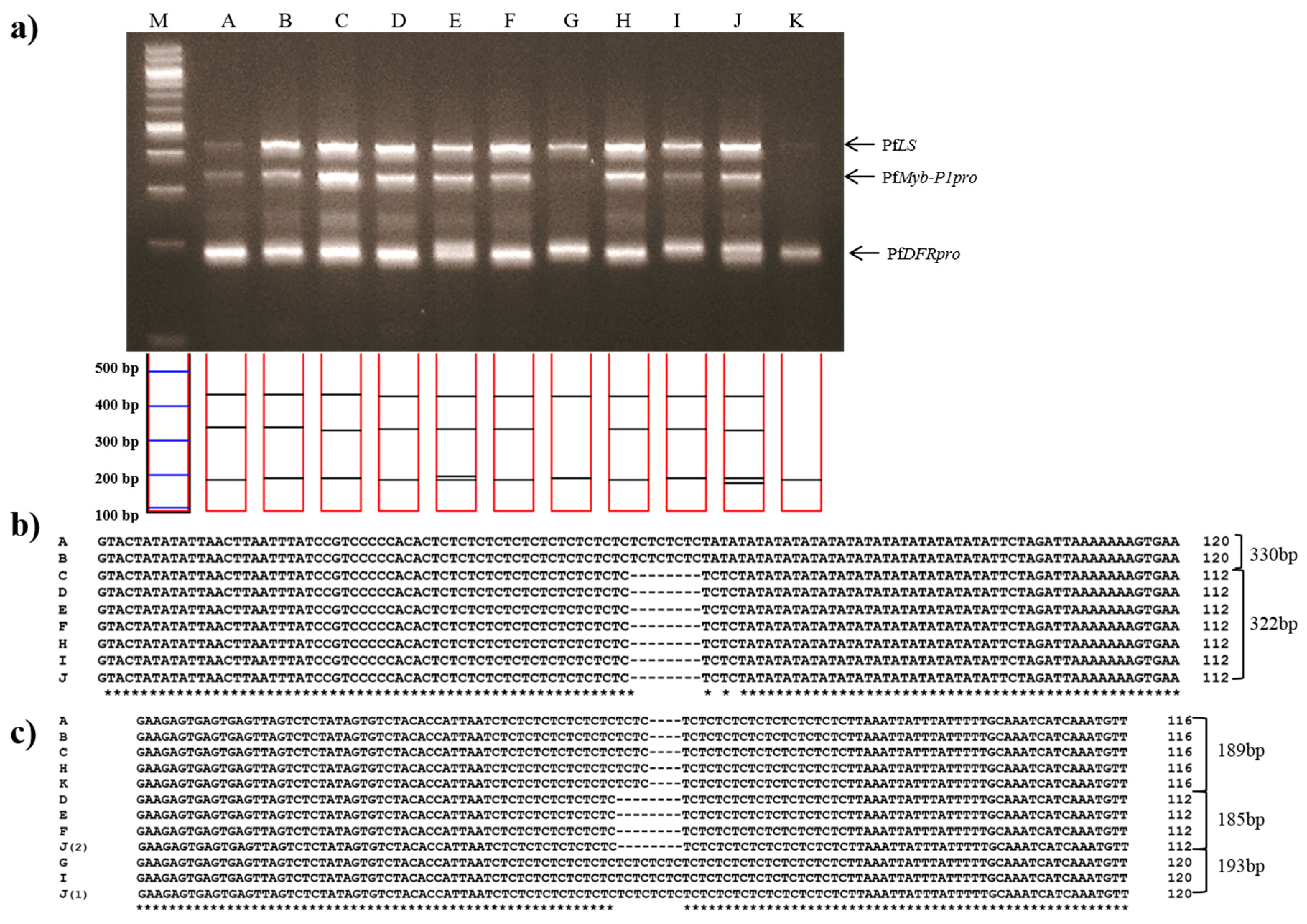
2. Results and Discussion
3. Materials and Methods
3.1. Plant Materials and Growth Conditions
3.2. Preparation of Genomic DNA
3.3. Gene Profiles and PCR Product Sequencing
3.4. Development of Specific Markers and Data Analysis
3.5. Development of the Multiplex-PCR Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lee, J.; Ohnishi, O. Genetic relationships among cultivated types of Perilla frutescens and their weedy types in East Asia revealed by AFLP markers. Gen. Res. Crop Evol. 2003, 50, 65–74. [Google Scholar] [CrossRef]
- Nitta, M.; Ohmi, O. Genetic relationships among two Perilla crops, shiso and egoma, and the weedy type revealed by RAPD markers. Genes Genet. Syst. 1999, 74, 43–48. [Google Scholar] [CrossRef]
- Müller-Waldeck, F.; Sitzmann, J.; Schnitzler, W.H.; Graßmann, J. Determination of toxic perilla ketone, secondary plant metabolites and antioxidative capacity in five Perilla frutescens L. varieties. Food Chem. Toxicol. 2010, 48, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Lozano, Y.F.; Gaydou, E.M.; Li, B. Antioxidant activities of polyphenols extracted from Perilla frutescens varieties. Molecules 2008, 14, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Park, K.H.; Lee, M.H.; Kim, H.T.; Seo, W.D.; Kim, J.Y.; Ha, T.J. Identification, characterisation, and quantification of phenolic compounds in the antioxidant activity-containing fraction from the seeds of Korean perilla (Perilla frutescens) cultivars. Food Chem. 2013, 136, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Chikako, Y.; Masatoshi, Y. Luteolin as an anti-inflammatory and anti-allergic constituent of Perilla frutescens. Biol. Pharm. Bull. 2002, 25, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, N.; Takano, H.; Sanbongi, C.; Yasuda, A.; Yanagisawa, R.; Inoue, K.I.; Yoshikawa, T. Anti-inflammatory and anti-allergic effect of rosmarinic acid (RA); inhibition of seasonal allergic rhinoconjunctivitis (SAR) and its mechanism. Biofactors 2004, 21, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Masatoshi, Y. Anti-inflammatory and anti-allergic actions by oral administration of a perilla leaf extract in mice. Biosci. Biotechnol. Biochem. 2001, 65, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Jeon, I.H.; Kim, H.S.; Kang, H.J.; Lee, H.S.; Jeong, S.I.; Kim, S.J.; Jang, S.I. Anti-inflammatory and antipruritic effects of luteolin from Perilla (P. frutescens L.) leaves. Molecules 2014, 19, 6941–6951. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Nakano, M.; Kawahata, T.; Otake, T.; Oishi, I.; Inami, R.; Nakanishi, T. Anti-HIV-1 activity of herbs in Labiatae. Biol. Pharm. Bull. 1998, 21, 829–833. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Tanaka, Y.; Fukuchi-Mizutani, M.; Fujiwara, H.; Fukui, Y.; Ashikari, T.; Kusumi, T. Molecular and biochemical characterization of a novel hydroxycinnamoyl-CoA: Anthocyanin 3-O-glucoside-6″-O-acyltransferase from Perilla frutescens. Plant Cell Physiol. 2000, 41, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Yamazaki, M.; Sugiyama, M.; Tanaka, Y.; Saito, K. Cloning and molecular analysis of structural genes involved in anthocyanin biosynthesis and expressed in a forma-specific manner in Perilla Frutescens. Plant Mol. Biol. 1997, 35, 915–927. [Google Scholar]
- He, Y.K.; Yao, Y.Y.; Chang, Y.N. Characterization of anthocyanins in Perilla frutescens var. acuta extract by advanced UPLC-ESI-IT-TOF-MSn method and their anticancer bioactivity. Molecules 2015, 20, 9155–9169. [Google Scholar] [PubMed]
- Yuba, A.; Honda, G.; Koezuka, Y.; Tabata, M. Genetic analysis of essential oil variants in Perilla frutescens. Biochem. Genet. 1995, 33, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Koezuka, Y.; Honda, G.; Tabata, M. Genetic control of phenylpropanoids in Perilla frutescens. Phytochemistry 1986, 25, 2085–2087. [Google Scholar] [CrossRef]
- Ohk, H.C.; Chae, Y.A. Analysis of volatile oil components and identification of chemotypes in Jaso (Perilla frutescens) collected in Korea. Korean J. Med. Crop Sci. 2004, 12, 97–101. [Google Scholar]
- Yamazaki, M.; Shibata, M.; Nishiyama, Y.; Springob, K.; Kitayama, M.; Shimada, N.; Aoki, T.; Ayabe, S.I.; Saito, K. Differential gene expression profiles of red and green forms of Perilla frutescens leading to comprehensive identification of anthocyanin biosynthetic genes. FEBS J. 2008, 275, 3494–3502. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Nakamura, M.; Suzuki, H.; Saito, K.; Yamazaki, M. High-throughput sequencing and de novo assembly of red and green forms of the Perilla frutescens var. crispa transcriptome. PLoS ONE 2015, 10, e0129154. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Kwon, S.J.; Lee, J.; Choi, I.Y.; Park, Y.J.; Choi, S.H.; Lee, J.K. Gene set by de novo assembly of Perilla species and expression profiling between P. frutescens (L.) var. frutescens and var. crispa. Gene 2015, 559, 155–163. [Google Scholar] [PubMed]
- Poczai, P.; Varga, I.; Laos, M.; Cseh, A.; Bell, N.; Valkonen, J.P.; Hyvönen, J. Advances in plant gene-targeted and functional markers: A review. Plant Method. 2013, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Kage, U.; Kumar, A.; Dhokane, D.; Karre, S.; Kushalappa, A.C. Functional molecular markers for crop improvement. Crit. Rev. Biotechnol. 2016, 36, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, M.; Makita, Y.; Springob, K.; Saito, K. Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var. crispa. Biochem. Eng. J. 2003, 14, 191–197. [Google Scholar] [CrossRef]
- Ohara, K.; Ujihara, T.; Endo, T.; Sato, F.; Yazaki, K. Limonene production in tobacco with Perilla limonene synthase cDNA. J. Exp. Bot. 2003, 54, 2635–2642. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.Z.; Yamagishi, E.; Yamazaki, M.; Saito, K. A constitutively expressed Myc-like gene involved in anthocyanin biosynthesis from Perilla frutescens: Molecular characterization, heterologous expression in transgenic plants and transactivation in yeast cells. Plant Mol. Biol. 1999, 41, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.Z.; Yamazaki, M.; Saito, K. A light inducible Myb-like gene that is specifically expressed in red Perilla frutescens and presumably acts as a determining factor of the anthocyanin forma. Mol. Gen. Genet. 1999, 262, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Merdinoglu, D.; Butterlin, G.; Bevilacqua, L.; Chiquet, V.; Adam-Blondon, A.F.; Decroocq, S. Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol. Breed. 2005, 5, 349–366. [Google Scholar] [CrossRef]
- James, D.; Schmidt, A.M.; Wall, E.; Green, M.; Masri, S. Reliable detection and identification of genetically modified maize, soybean, and canola by multiplex PCR analysis. J. Agric. Food Chem. 2003, 51, 5829–5834. [Google Scholar] [CrossRef] [PubMed]
- Group, C.P.W.; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Korea Food and Drug Administration. The Korean Herbal Pharmacopoeia, 9th ed.; KFDA: Soul, Korea, 2008; Volume 255, pp. 107–1088. [Google Scholar]
- Koichiro, T.; Glen, S.; Daniel, P.; Alan, F.; Sudhir, K. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.G.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; Thompson, J.D.; Higgins, D. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Shete, S.; Tiwari, H.; Elston, R.C. On estimating the heterozygosity and polymorphism information content value. Theor. Popul. Biol. 2000, 57, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples are available from the authors. |


| No. | Scientific Name | Common Name | Collection | Location |
|---|---|---|---|---|
| 1 | P. frutescens var. japonica | Cham-Dlggae | Yongin-si, Gyeonggi-do, Korea | 37°06′32.7″N 127°07′09.7″E |
| 2 | Ip-Dlggae | Yongin-si, Gyeonggi-do, Korea | 37°06′32.7″N 127°07′09.7″E | |
| 3 | Bora-Dlggae | Hwasun-gun, Jeollanam-do, Korea | 35°02′47.5″N 126°57′13.9″E | |
| 4 | P. frutescens var. crispa | Jureum-soyeop | Hwasun-gun, Jeollanam-do, Korea | 35°02′47.5″N 126°57′13.9″E |
| 5 | Jangheung-gun, Jeollanam-do, Korea | 34°40′03.0″N 126°56′44.6″E | ||
| 6 | Yongin-si, Gyeonggi-do, Korea | 37°06′32.7″N 127°07′09.7″E | ||
| 7 | P. frutescens var. acuta | Jasoyeop | Yecheon-gun, Gyeongsangbuk-do, Korea | 35°59′28.1″N 128°55′33.3″E |
| 8 | Namyangju-si, Gyeonggi-do, Korea | 37°38′58.1″N 127°11′31.7″E | ||
| 9 | Hwasun-gun, Jeollanam-do, Korea | 35°02′47.5″N 126°57′13.9″E | ||
| 10 | P. frutescens f. viridis | Chungsoyeop | Yecheon-gun, Gyeongsangbuk-do, Korea | 35°59′28.1″N 128°55′33.3″E |
| 11 | Namyangju-si, Gyeonggi-do, Korea | 37°38′58.1″N 127°11′31.7″E | ||
| 12 | Hwasun-gun, Jeollanam-do, Korea | 35°02′47.5″N 126°57′13.9″E |
| Gene Name | Genbank Accession | Primer Sequences 5′ to 3′ | Tm °C | Size (bp) | Location |
|---|---|---|---|---|---|
| LS1 a | AF241791 | F:GAATGTGCATTTGGAATATTATAAG R:TTGTGAAGCGTACGTTCTGCCGGAG | 58 | 255 | 5′UTR |
| LS2 a | F:CATCAGACGGTCTAACGGGTAACAA R1:CACTATGCTCTATAATTAATTGGGTG R2: AAATATATACCATAAAATGGATTAG R3:GTTTTGTCCGACACTAACGTGTGAG | 60 59 61 58 | 253 515 576 | Exon1 to Intron1 | |
| LS3 a | F: GAATAGTTAATTAATGGGTGTGCAG R:AGGAGTCTGAATGCAAGAGCTGTCG | 57 | 308 | Exon2 | |
| LS4 a | F: CCTGGTAGATACATATTTATATTAT R:TTTAGCACAGTGCACGAGCCACACG | 60 | 189 | Intron2 | |
| LS5 a | F:CAACACTTGAACTGTGGCGGGTTGA R:ATGTCCAAAGAGTGGCGCACACATG | 58 | 358 | Exon3 | |
| LS6 a | F: CTGGTGGATTGATGCCTATAAGAGG R: TACATACATACATATATAACATGAG | 59 | 223 | Exon3 to Intron4 | |
| LS7 a | F: GCTATTCACCGAGGCGATTCGGAGG R1: ATTTTAATTTGATAGGCTGATTGAG R2:TGTCGTCCACGAAATCGTTGAGTGC | 60 60 | 301 430 | Exon4 to 3′UTR | |
| LS8 a | F: AGCGGCCCTGTAATGCTTTGCCATG R1:CACATCGCCTCGACTCACCTCCTCC R2:ATAAGCCATCTGATGTGGCGTCGCG | 62 59 | 284 492 | 3′UTR | |
| Myb-P1pro1 b | AB024053 | F: GAATTGGAAACAAATTAAGGATCGG R: TGATGTCTGGCCTCAGGTAGTTCAT | 59 | 612 | 5′UTR |
| Myb-P1pro2 b | F: TTAAGGATCGGAGATCGAATGAGG R: AGCTTGATCATCAGCTCTTCTTCAT | 60 | 657 | 5′UTR | |
| Myb-P1pro3 b | F: ATGAACTACCTGAGGCCAGACATCA R: ATGAGTATGAATATTTCTTTGAGGT | 58 | 601 | Coding | |
| DFRpro1 c | AB024052 | F: GAATTCGAGCTCGAATATGTACATT R: CGACCAACTATTATAGGCATAGCTCC | 59 | 745 | 5′UTR |
| DFRpro2 c | F: GGAGCTATGCCTATAATAGTTGGTCG R: GAAACAATCTTGACCACCATTCTTG | 60 | 619 | 5′UTR |
| Marker Name | Marker Sequence | Product Size (bp) | Number of Alleles | PIC | ||||
|---|---|---|---|---|---|---|---|---|
| J a | A b | C c | V d | |||||
| PfLS | F | GCTATTCACCGAGGCGATTCGGAGG | N/A | 430 | 430 | 430 | N/A | N/A |
| R | TGTCGTCCACGAAATCGTTGAGTGC | |||||||
| PfMybpro | F | GTACTATATATTAACTTAATTTATCCGT | 314 316 | 330 | 322 | 315 | 5 | 0.23 |
| R | GCTTGATCATCAGCTCTTCTTCATCCAC | |||||||
| PfDFRpro | F | GAAGAGTGAGTGAGTTAGTCTCTATAGT | 189 202 | 187 189 | 185 189 | 193 | 7 | 0.36 |
| R | GCAAGAGATTTATGAGCATGTTCTAGAT | |||||||
| Herbal Makers | Material Component | Source |
|---|---|---|
| A | Dried leaves and twigs of Jasoyeop or Jureum-soyeop products | Gapyeong-gun, Gyeonggi-do, Korea |
| B | Andong-si, Gyeongsangbuk-do, Korea | |
| C | Yeongcheon-si, Gyeongsangbuk-do, Korea | |
| D | Jecheon-si, Chungcheongbuk-do, Korea | |
| E | Gapyeong-gun, Gyeonggi-do, Korea | |
| F | Sancheong-gun, Gyeongsangnam-do, Korea | |
| G | Taean-gun, Chungcheongnam-do, Korea | |
| H | Yeongcheon-si, Gyeongsangbuk-do, Korea | |
| I | Uiseong-gun, Gyeongsangbuk-do, Korea | |
| J | Hunan Province, China | |
| K | Yeongcheon-si, Gyeongsangbuk-do, Korea |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Kim, A.-Y.; Jo, A.; Choi, H.; Cho, S.-S.; Choi, C. Development of User-Friendly Method to Distinguish Subspecies of the Korean Medicinal Herb Perilla frutescens Using Multiplex-PCR. Molecules 2017, 22, 665. https://doi.org/10.3390/molecules22040665
Kim Y, Kim A-Y, Jo A, Choi H, Cho S-S, Choi C. Development of User-Friendly Method to Distinguish Subspecies of the Korean Medicinal Herb Perilla frutescens Using Multiplex-PCR. Molecules. 2017; 22(4):665. https://doi.org/10.3390/molecules22040665
Chicago/Turabian StyleKim, Yonguk, Ah-Young Kim, Ara Jo, Hakjoon Choi, Seung-Sik Cho, and Chulyung Choi. 2017. "Development of User-Friendly Method to Distinguish Subspecies of the Korean Medicinal Herb Perilla frutescens Using Multiplex-PCR" Molecules 22, no. 4: 665. https://doi.org/10.3390/molecules22040665






