Application of UHPLC-ESI-Q-TOF-MS to Identify Multiple Constituents in Processed Products of the Herbal Medicine Ligustri Lucidi Fructus
Abstract
:1. Introduction
2. Results and Discussion
2.1. UHPLC-ESI-Q-TOF-MS Analysis of Constituents in Processed-LLF
2.2. Identification of Phenylethanoids and Glycosides 2, 3, 7, 9, 13, 14, 23, 26
2.3. Identification of Flavonoids 16, 18, 20, 24, 28, 33, 39
2.4. Identification of Iridoids 4−6, 8, 10−12, 15, 17, 19, 21, 22, 25, 27, 29−32, 34−38, 40, 41
2.5. Identification of Triterpenoids 42−50
2.6. Other Compounds
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Plant Materials and Processing
3.3. Sample and Standard Solution Preparation
3.4. UHPLC-ESI-QTOF-MS System and Conditions
3.5. Data Processing
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gu, G. Shen Nong’s Herbal Classic; Huaxia Publishing House: Beijing, China, 1999; p. 43. [Google Scholar]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2015; Volume I, p. 45. [Google Scholar]
- Gao, L.; Li, C.; Wang, Z.; Liu, X.; You, Y.; Wei, H.; Guo, T. Ligustri lucidi fructus as a traditional Chinese medicine: A review of its phytochemistry and pharmacology. Nat. Prod. Res. 2015, 29, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Zhan, F.; Zhang, X.; Jiang, H.; Zhao, Z. Assay of Ligustrum lucidum Ait. with and without steaming with wine by HPLC-ESI/MS. Chin. Tradit. Pat. Med. 2013, 35, 2707–2710. [Google Scholar]
- Han, L.; Boakye-Yiadom, M.; Liu, E.; Zhang, Y.; Li, W.; Song, X.; Fu, F.; Gao, X. Structural characterisation and identification of phenylethanoid glycosides from Cistanches deserticola YC Ma by UHPLC/ESI-QTOF-MS/MS. Phytochem. Anal. 2012, 23, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Amessis-Ouchemoukh, N.; Abu-Reidah, I.M.; Quirantes-Piné, R.; Rodríguez-Pérez, C.; Madani, K.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Tentative characterisation of iridoids, phenylethanoid glycosides and flavonoid derivatives from Globularia alypum L. (Globulariaceae) leaves by LC-ESI-QTOF-MS. Phytochem. Anal. 2014, 25, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.H.; Tong, X.; Wang, J.X.; Zou, W.; Cao, H.; Su, W.W. Rapid separation and identification of multiple constituents in traditional Chinese medicine formula Shenqi Fuzheng Injection by ultra-fast liquid chromatography combined with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2013, 74, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Quirantes-Piné, R.; Lozano-Sánchez, J.; Herrero, M.; Ibáñez, E.; Segura-Carretero, A.; Fernández-Gutiérrez, A. HPLC-ESI-QTOF-MS as a powerful analytical tool for characterizing phenolic compounds in olive leaf extracts. Phytochem. Anal. 2013, 24, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.T.; Wu, X.; Rui, W.; Guo, J.; Feng, Y.F. UPLC/Q-TOF-MS analysis for identification of hydrophilic phenolics and lipophilic diterpenoids from Radix Salviae Miltiorrhizae. Acta Chromatogr. 2015, 27, 711–728. [Google Scholar] [CrossRef]
- Song, J.; Zhao, L.; Rui, W.; Guo, J.; Feng, Y. Identification and Fragmentation Pattern Analysis of Iridoid Glycosides Fromfructus Ligustri Lucidiby UPLC/ESI-QTOF-MS. J. Liq. Chromatogr. Relat. Technol. 2014, 37, 1763–1770. [Google Scholar] [CrossRef]
- Peralbo-Molina, A.; Priego-Capote, F.; Luque de Castro, M.D. Tentative identification of phenolic compounds in olive pomace extracts using liquid chromatography-tandem mass spectrometry with a quadrupole-quadrupole-time-of-flight mass detector. J. Agric. Food. Chem. 2012, 60, 11542–11550. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yin, Z.; Ye, W.; Shen, W. Chemical constituents from fruits of Ligustrum lucidum. China J. Chin. Mater. Med. 2010, 35, 861–864. [Google Scholar]
- Klen, T.J.; Wondra, A.G.; Vrhovsek, U.; Vodopivec, B.M. Phenolic profiling of olives and olive oil process-derived matrices using UPLC-DAD-ESI-QTOF-HRMS analysis. J. Agric. Food. Chem. 2015, 63, 3859–3872. [Google Scholar] [CrossRef] [PubMed]
- Taamalli, A.; Arraez-Roman, D.; Ibanez, E.; Zarrouk, M.; Segura-Carretero, A.; Fernandez-Gutierrez, A. Optimization of microwave-assisted extraction for the characterization of olive leaf phenolic compounds by using HPLC-ESI-TOF-MS/IT-MS(2). J. Agric. Food. Chem. 2012, 60, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Ola, S.S.; Catia, G.; Marzia, I.; Vincieri, F.F.; Akindahunsi, A.A.; Mulinacci, N. HPLC/DAD/MS characterisation and analysis of flavonoids and cynnamoil derivatives in four Nigerian green-leafy vegetables. Food Chem. 2009, 115, 1568–1574. [Google Scholar] [CrossRef]
- Cuyckens, F.; Rozenberg, R.; de Hoffmann, E.; Claeys, M. Structure characterization of flavonoid O-diglycosides by positive and negative nano-electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 2001, 36, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Hvattum, E.; Ekeberg, D. Study of the collision-induced radical cleavage of flavonoid glycosides using negative electrospray ionization tandem quadrupole mass spectrometry. J. Mass Spectrom. 2003, 38, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Arraez-Roman, D.; Segura-Carretero, A.; Menendez, J.A.; Menendez-Gutierrez, M.P.; Micol, V.; Fernandez-Gutierrez, A. Qualitative screening of phenolic compounds in olive leaf extracts by hyphenated liquid chromatography and preliminary evaluation of cytotoxic activity against human breast cancer cells. Anal. Bioanal. Chem. 2010, 397, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, M.; Lin, Z.; Jiang, H.; Niu, Y.; Wang, H.; Chen, S. Comprehensive identification of 125 multifarious constituents in Shuang-huang-lian powder injection by HPLC-DAD-ESI-IT-TOF-MS. J. Pharm. Biomed. Anal. 2015, 115, 86–106. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Xiang, T.; Hou, B.; Liang, W.; Yin, S.; Zhou, X. Chemical constituents from fruits of Ligustrum lucidum. Acta Bot. Sin. 1997, 40, 83–87. [Google Scholar]
- Aoki, S.; Honda, Y.; Kikuchi, T.; Miura, T.; Sugawara, R.; Yaoita, Y.; Kikuchi, M.; Machida, K. Six New Secoiridoids from the Dried Fruits of Ligustrum lucidum. Chem. Pharm. Bull. 2012, 60, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Buiarelli, F.; Cartoni, G.; Coccioli, F.; Jasionowska, R.; Margherita, P. Analysis by liquid chromatography-tandem mass spectrometry of biophenolic compounds in olives and vegetation waters, Part I. J. Sep. Sci. 2003, 26, 409–416. [Google Scholar] [CrossRef]
- Tóth, G.; Alberti, Á.; Sólyomváry, A.; Barabás, C.; Boldizsár, I.; Noszál, B. Phenolic profiling of various olive bark-types and leaves: HPLC-ESI/MS study. Ind. Crops Prod. 2015, 67, 432–438. [Google Scholar] [CrossRef]
- Rubio-Senent, F.; Lama-Munoz, A.; Rodriguez-Gutierrez, G.; Fernandez-Bolanos, J. Isolation and identification of phenolic glucosides from thermally treated olive oil byproducts. J. Agric. Food. Chem. 2013, 61, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Yu, Y.; Ablajan, K.; Li, L.; Fan, B.; Peng, J.; Yan, H.; Ma, F.; Nie, Y. Seasonal variations in metabolite profiling of the fruits of Ligustrum lucidum Ait. Rapid Commun. Mass Spectrom. 2011, 25, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Tóth, G.; Barabás, C.; Tóth, A.; Kéry, Á.; Béni, S.; Boldizsár, I.; Varga, E.; Noszál, B. Characterization of antioxidant phenolics in Syringa vulgaris L. flowers and fruits by HPLC-DAD-ESI-MS. Biomed. Chromatogr. 2016, 30, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Villalba, R.; Carrasco-Pancorbo, A.; Oliveras-Ferraros, C.; Vazquez-Martin, A.; Menendez, J.A.; Segura-Carretero, A.; Fernandez-Gutierrez, A. Characterization and quantification of phenolic compounds of extra-virgin olive oils with anticancer properties by a rapid and resolutive LC-ESI-TOF MS method. J. Pharm. Biomed. Anal. 2010, 51, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, P.; Termentzi, A.; Michel, T.; Gikas, E.; Halabalaki, M.; Skaltsounis, A.L. From olive drupes to olive oil. An HPLC-orbitrap-based qualitative and quantitative exploration of olive key metabolites. Planta Med. 2013, 79, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Savarese, M.; Demarco, E.; Sacchi, R. Characterization of phenolic extracts from olives (Olea europaea cv. Pisciottana) by electrospray ionization mass spectrometry. Food Chem. 2007, 105, 761–770. [Google Scholar] [CrossRef]
- Garcia-Villalba, R.; Tomas-Barberan, F.A.; Fanca-Berthon, P.; Roller, M.; Zafrilla, P.; Issaly, N.; Garcia-Conesa, M.T. Targeted and untargeted metabolomics to explore the bioavailability of the secoiridoids from a seed/fruit extract (Fraxinus angustifolia Vahl) in human healthy volunteers: A preliminary study. Molecules 2015, 20, 22202–22219. [Google Scholar] [CrossRef] [PubMed]
- Michel, T.; Khlif, I.; Kanakis, P.; Termentzi, A.; Allouche, N.; Halabalaki, M.; Skaltsounis, A.-L. UHPLC-DAD-FLD and UHPLC-HRMS/MS based metabolic profiling and characterization of different Olea europaea organs of Koroneiki and Chetoui varieties. Phytochem. Lett. 2015, 11, 424–439. [Google Scholar] [CrossRef]
- Fu, S.; Arráez-Román, D.; Menendez, J.A.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Characterization of isomers of oleuropein aglycon in olive oils by rapid-resolution liquid chromatography coupled to electrospray time-of-flight and ion trap tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tu, F.; Dai, Y.; Yao, X. Chemical Constituents of Ligustrum lucidum. China Pharm. 2011, 22, 2931–2933. [Google Scholar]
- Feng, J.; Feng, Z.; Wang, J.; Cui, Y. Study on the Triterpenoids from the Fruits of Ligustrum lucidum. J. Chin. Med. Mater. 2011, 34, 1540–1544. [Google Scholar]
- Chen, Q.; Yang, L.; Zhang, G.; Wang, F. Bioactivity-guided isolation of antiosteoporotic compounds from Ligustrum lucidum. Phytother. Res. 2013, 27, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.Y.; Chen, S.C.; Wu, K.J.; Kuo, H.C.; Hseu, Y.C.; Ching, H.; Wu, C.R. Antioxidant phenolic profile from ethyl acetate fraction of Fructus Ligustri Lucidi with protection against hydrogen peroxide-induced oxidative damage in SH-SY5Y cells. Food Chem. Toxicol. 2012, 50, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yang, N.; Qian, S.; Xie, N.; Yu, M.; Duan, J. Stduy on flavonoids in Ligustrum lucidum. China J. Chin. Mater. Med. 2007, 30, 538–540. [Google Scholar]
- Long, F.; Deng, L.; Chen, Y. Study on the chemical constituents in the flowers of Ligustrum lucium. West China J. Pharm. Sci. 2011, 26, 97–100. [Google Scholar]
- Nagy, M.; Krizkova, L.; Mucaji, P.; Kontsekova, Z.; Sersen, F.; Krajcovic, J. Antimutagenic activity and radical scavenging activity of water infusions and phenolics from ligustrum plants leaves. Molecules 2009, 14, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.R.; Franzyk, H.; Wallander, E. Chemotaxonomy of the Oleaceae: iridoids as taxonomic markers. Phytochemistry 2002, 60, 213–231. [Google Scholar] [CrossRef]
- Obied, H.K.; Bedgood, D.R., Jr.; Prenzler, P.D.; Robards, K. Chemical screening of olive biophenol extracts by hyphenated liquid chromatography. Anal. Chim. Acta 2007, 603, 176–189. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Dong, H.; Xu, H.; Ye, W.; Sun, H.; But, P.P.-H. Secoiridoid constituents from the fruits of Ligustrum lucidum. Phytochemistry 2001, 56, 327–330. [Google Scholar] [CrossRef]
- He, Z.; But, P.P.-H.; Chan, T.-W.D.; Dong, H.; Xu, H.; Lau, C.; Sun, H. Antioxidative Glucosides from the Fruits of Ligustrum lucidum. Chem. Pharm. Bull. 2001, 49, 780–784. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Gomes, L.; Leitão, F.; Bronze, M.; Coelho, A.V.; Boas, L.V. Secoiridoids in olive seed: characterization of nüzhenide and 11-methyl oleosides by liquid chromatography with diode array and mass spectrometry. Grasas Aceites 2010, 61, 157–164. [Google Scholar] [CrossRef]
- Jiang, Q.; Jiang, H.; Li, H.; Zhang, X. Contents dynamic changes of four secoiridoid glycosides under steaming time spans with wine in Ligubtrum lucidum Ait. Chin. Tradit. Pat. Med. 2014, 36, 2561–2564. [Google Scholar]
- Huang, X.J.; Wang, Y.; Yin, Z.Q.; Ye, W.C. Two new dimeric secoiridoid glycosides from the fruits of Ligustrum lucidum. J. Asian Nat. Prod. Res. 2010, 12, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, X.; Li, H.; Cui, W. Dynamic changes of neonuezhenide, oleoside-11-methyl ester and nuezhenidic acid in wine steaming process of Ligustri Lucidi Fructus. Chin. J. Exp. Tradit. Med. Formulae 2014, 20, 14–17. [Google Scholar]
- Yim, T.K.; Wu, W.K.; Pak, W.F.; Ko, K.M. Hepatoprotective action of an oleanolic acid-enriched extract of Ligustrum lucidum fruits is mediated through an enhancement on hepatic glutathione regeneration capacity in mice. Phytother. Res. 2001, 15, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, C.; Ren, R.; Liu, R. Simultaneous determination of seven major triterpenoids in Pyrola decorata H. Andres by LC-MS method. Pharmazie 2012, 67, 822–826. [Google Scholar]
- Masullo, M.; Montoro, P.; Autore, G.; Marzocco, S.; Pizza, C.; Piacente, S. Quali-quantitative determination of triterpenic acids of Ziziphus jujuba fruits and evaluation of their capability to interfere in macrophages activation inhibiting NO release and iNOS expression. Food Res. Int. 2015, 77, 109–117. [Google Scholar] [CrossRef]
- Li, H.; Liu, Q.; Zhang, L.; Yao, W.; Ding, A. Optimization of processing technology of Ligustri Lucidi Fructus stewed with wine based on analytic hierarchy process and multi-index orthogonal test. Chin. Tradit. Herb. Drugs 2016, 47, 2832–2837. [Google Scholar]
Sample Availability: Samples of all the compounds are not available from the authors. |
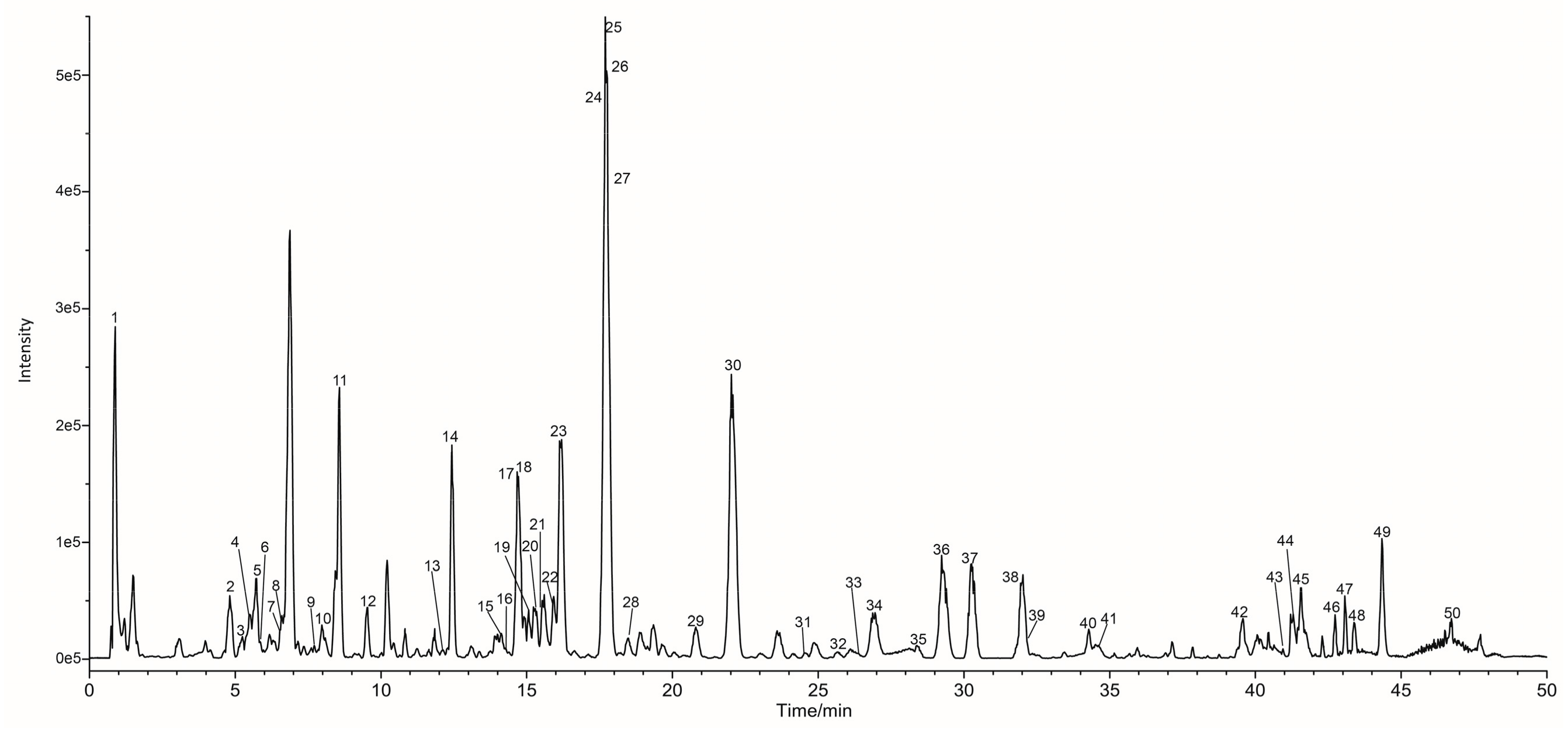
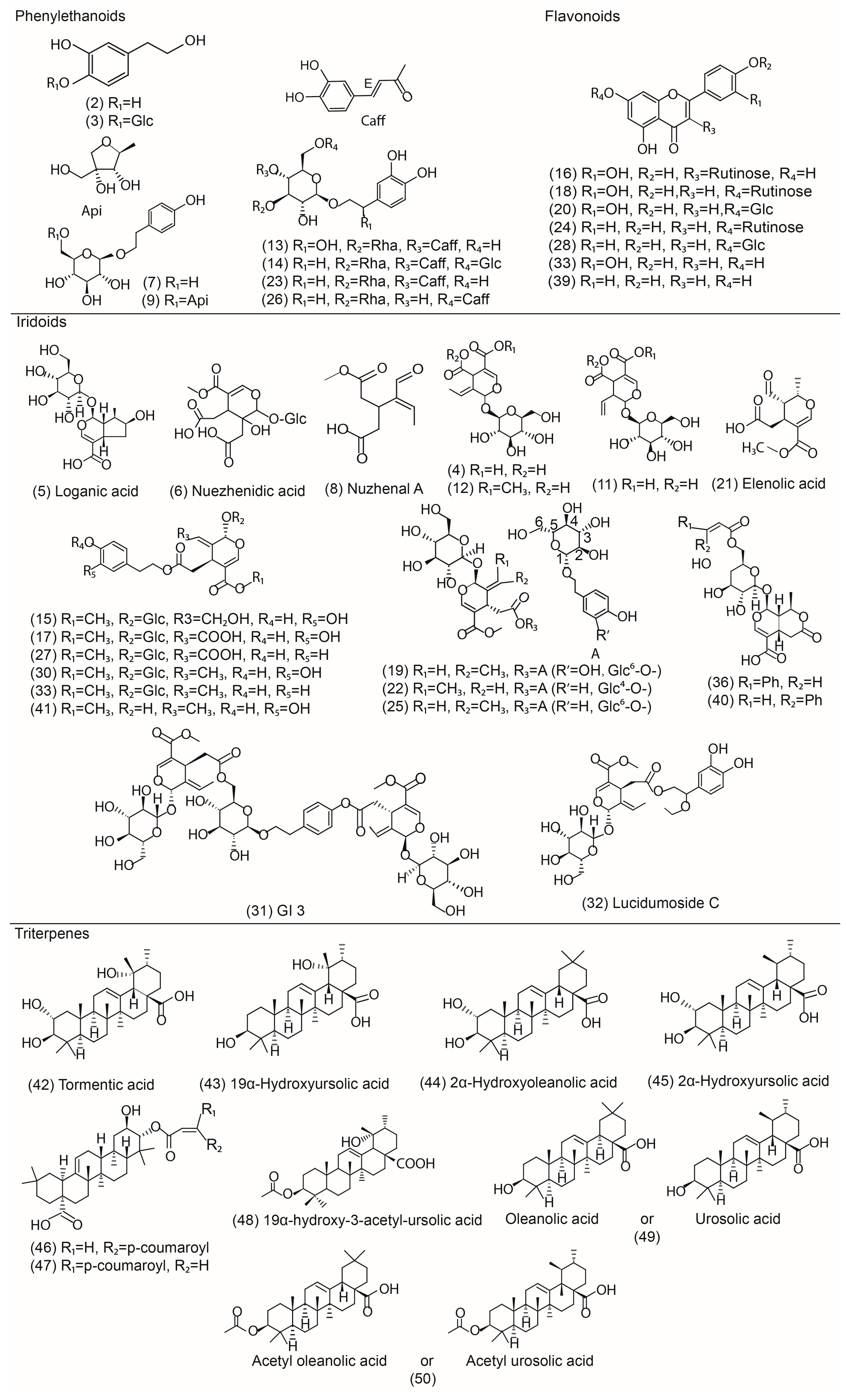

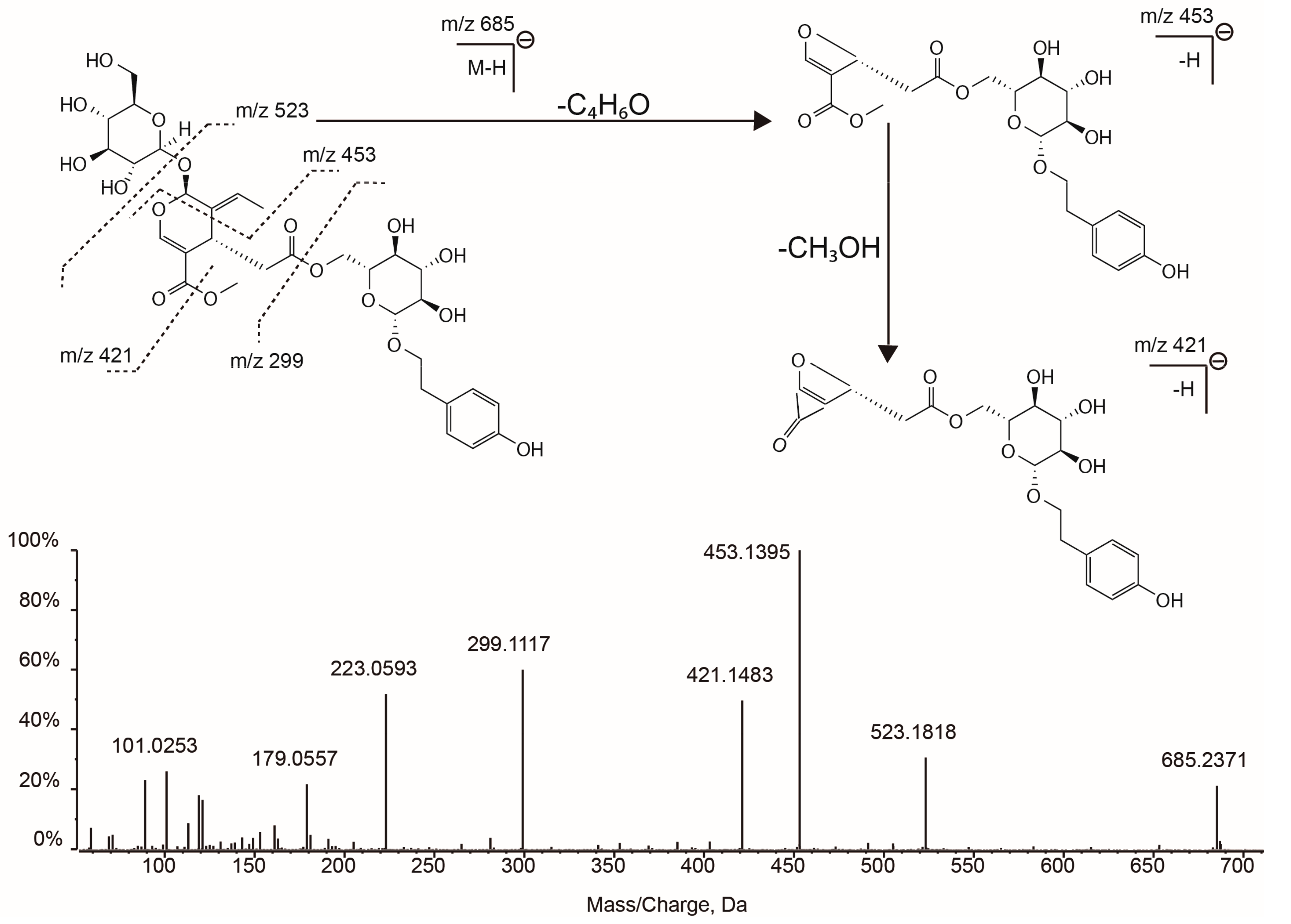
| Peak | RT (min) | Formula | [M − H] − | MS2 Fragments (Relative Abundance) | Proposed Compound | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| m/z theo | m/z exp | Error (ppm) | ||||||
| Phenylethanoids | ||||||||
| 2 | 4.81 | C8H10O3 | 153.0557 | 153.0556 | −1 | 123.0456 (100) | Hydroxytyrosol | [11] |
| 3 | 5.22 | C14H20O8 | 315.1085 | 315.1089 | 1.1 | 153.0554 (94.24), 123.0454 (100) | Hydroxytyrosol Glucoside | [11] |
| 7 | 6.60 | C14H20O7 | 299.1136 | 299.1140 | 1.2 | 137.0587 (13.60), 119.0500 (83.73), 113.0250 (27.20), 101.0270 (24.53), 59.0194 (100) | Salidroside | standard |
| 9 | 7.72 | C19H28O11 | 431.1559 | 431.1558 | −0.1 | 299.1114 (10.82), 149.0457 (25.15), 119.0486 (12.54), 101.0255 (12.87) | Osmanthuside H | [12] |
| 13 | 12.13 | C29H36O16 | 639.1931 | 639.1933 | 0.4 | 621.1878 (47.63), 459.1506 (10.31), 179.0341 (69.58), 161.0238 (100), 135.0447 (12.63) | β-Hydroxyverbascoside | [13] |
| 14 | 12.44 | C35H46O20 | 785.2510 | 785.2517 | 0.9 | 623.2239 (39.09), 461.1683 (2.63), 179.0371 (2.79), 161.0236 (35.63), 135.0450 (3.32) | Echinacoside | [5] |
| 23 | 16.18 | C29H36O15 | 623.1981 | 623.1989 | 1.3 | 461.1669 (35.12), 161.0234 (100), 179.0342 (7.52), 135.0447 (13.66) | Verbascoside | standard |
| 26 | 17.81 | C29H36O15 | 623.1981 | 623.1983 | 0.3 | 461.1679 (34.39), 161.0235 (100), 179.0340 (6.70), 135.0448 (12.00) | Isoverbascoside | [5] |
| Flavonoids | ||||||||
| 16 | 14.39 | C27H30O16 | 609.1461 | 609.1466 | 0.8 | 301.0356 (39.90), 300.0278 (89.95), 178.9991 (4.59), 151.0033 (4.34) | Quercetin-3-O-rutinoside | [8,14] |
| 18 | 14.78 | C27H30O15 | 593.1512 | 593.1511 | −0.2 | 285.0407 (100), 284.0327 (9.31) | Luteolin-7-O-rutinoside | [15] |
| 20 | 15.28 | C21H20O11 | 447.0933 | 447.0934 | 0.2 | 285.0397 (100), 284.0320 (44.59) | Luteolin-7-O-glucoside | standard |
| 24 | 17.60 | C27H30O14 | 577.1563 | 577.1557 | −1 | 311.0559 (0.57), 269.0450 (100) | Apigenin-7-O-rutinoside | [16] |
| 28 | 18.48 | C21H20O10 | 431.0984 | 431.0983 | −0.1 | 269.0454 (26.47), 268.0369 (100) | Apigenin-7-O-glucoside | [17] |
| 33 | 26.33 | C15H10O6 | 285.0405 | 285.0408 | 1.2 | 151.0033 (14.30), 133.0297 (68.21) | Luteolin | standard |
| 39 | 32.14 | C15H10O5 | 269.0456 | 269.0455 | −0.2 | 151.0041 (14.36), 117.0353 (58.84) | Apigenin | standard |
| Iridoids | ||||||||
| 4 | 5.50 | C16H22O11 | 389.1089 | 389.1093 | 0.9 | 345.1190 (3.03), 227.0557 (37.80), 209.0454 (14.54), 183.0660 (87.28), 165.0551 (67.05), 121.0665 (100) | Oleoside | [13,18] |
| 5 | 5.71 | C16H24O10 | 375.1297 | 375.1300 | 0.9 | 331.1405 (70.23), 169.0872 (3.45), 151.0761 (25.68), 213.0783 (1.16), 113.0250 (39.00) | Loganic acid | [11,19] |
| 6 | 5.84 | C17H24O14 | 451.1093 | 451.1092 | −0.3 | 433.0995 (11.52), 271.0457 (22.27), 239.0177 (14.00), 227.0544 (28.26), 195.0296 (70.07), 183.0653 (25.05), 151.0398 (100), 123.0459 (35.06) | Nuezhenidic acid | [20] |
| 8 | 6.61 | C10H14O5 | 213.0768 | 213.0777 | 4.1 | 183.0666 (100), 151.0768 (54.45), 121.0670 (31.32), 107.0889 (9.07) | Nuzhenal A | [21] |
| 10 | 8.06 | C17H24O11 | 403.1246 | 403.1247 | 0.3 | 223.0598 (54.55), 179.0555 (26.50), 121.0307 (41.08), 119.0355 (57.87), 113.0242 (34.44), 101.0249 (55.77), 89.0264 (97.55), 59.0190 (100) | Oleoside 11-methyl ester (isomer) | [8,11,22] |
| 11 | 8.57 | C16H22O11 | 389.1089 | 389.1093 | 0.9 | 345.1192 (29.00), 209.0449 (17.65), 183.0658 (26.15), 165.0552 (41.48), 121.0656 (62.61) | Secologanoside | [13,18,19] |
| 12 | 9.53 | C17H24O11 | 403.1246 | 403.1250 | 1 | 223.0608 (47.43), 179.0564 (26.79), 121.0290 (25.08), 119.0360 (52.49), 113.0251 (30.06), 101.0266 (56.07), 89.0270 (100), 59.0193 (83.41) | Oleoside 11-methyl ester | [8,11,22] |
| 15 | 14.12 | C25H32O14 | 555.1719 | 555.1725 | 1 | 323.0771(3.64), 223.0603(7.70), 151.0397(100), 123.0453(11.68) | 10-Hydroxyoleuropein | [8,23] |
| 17 | 14.71 | C25H30O15 | 569.1512 | 569.1517 | 0.9 | 389.0899 (30.33), 209.0452 (46.58), 151.0402 (100), 123.0455 (26.25) | Oleuropeinic acid | [24,25] |
| 19 | 15.07 | C31H42O18 | 701.2298 | 701.2311 | 1.8 | 539.1797 (12.19), 469.1375 (20.41), 437.1443 (10.17), 315.1081 (100) | Neonuzhenide | [26] |
| 21 | 15.48 | C11H14O6 | 241.0718 | 241.0724 | 2.5 | 139.0035 (100), 127.0403 (54.59), 121.0295 (27.57), 101.0262 (44.93), 95.0522 (61.86) | Elenolic acid | [27,28] |
| 22 | 15.94 | C31H42O17 | 685.2349 | 685.2360 | 1.6 | 523.1853 (75.34), 453.1422 (98.87), 421.1519 (53.10), 299.1134 (100), 223.0606 (82.17), 179.0558 (29.48), 119.0371 (31.24) | Nuezhenide (isomer) | -- |
| 25 | 17.72 | C31H42O17 | 685.2349 | 685.2356 | 0.9 | 523.1818 (30.54), 453.1395 (100),421.1483 (49.59), 299.1117 (59.94), 223.0593 (51.80), 179.0557 (21.62), 119.0374 (17.98) | Specnuezhenide | standard |
| 27 | 17.83 | C25H30O14 | 553.1563 | 553.1558 | −0.8 | 509.1670 (11.95), 477.1435 (4.16), 391.1015 (8.35), 373.0941 (41.57), 347.1143 (100), 209.0447 (82.67) | Ligustrosidic acid | [10,25] |
| 29 | 20.80 | C31H42O17 | 685.2349 | 685.2356 | 0.9 | 523.1856 (24.96), 453.1429 (44.64), 421.1518 (22.12), 385.1150 (22.09), 299.1134 (100), 223.0609 (24.93), 179.0555 (20.58), 119.0364 (25.63) | Isonuezhenide | [26] |
| 30 | 22.06 | C25H32O13 | 539.1770 | 539.1773 | 0.5 | 403.1253 (14.23), 377.1241 (57.51), 307.0813 (91.75), 275.0877 (79.24), 223.0601 (46.21), 179.0563 (19.28), 149.0241 (100), 139.0381 (54.09) | Oleuropein | [8,11,29] |
| 31 | 24.56 | C48H64O27 | 1071.3562 | 1071.3581 | 1.7 | 909.3186 (10.15), 839.2691 (9.04), 771.2401 (31.56), 685.2395 (36.40), 523.1839 (26.89), 453.1413 (29.53), 403.1255 (18.24), 385.1166 (12.15), 299.1139 (11.67), 223.0605 (23.17) | G 13 | [13,30] |
| 32 | 25.68 | C27H36O14 | 583.2032 | 583.2035 | 0.5 | 537.1649 (31.84), 403.1255 (35.57), 223.0603 (32.74), 151.0401 (100) | Lucidumoside C | [8] |
| 34 | 26.91 | C25H32O12 | 523.1821 | 523.1820 | −0.2 | 361.1296 (27.22), 291.0870 (100), 259.0968 (22.10), 101.0260 (20.27) | Ligustroside | [8,10,31] |
| 35 | 28.41 | C48H64O27 | 1071.3562 | 1071.3585 | 2.1 | 909.3161 (22.61), 839.2736 (16.57), 771.2440 (20.00), 685.2429 (45.75), 523.1855 (35.60), 453.1438 (23.63), 403.1267 (14.76), 385.1145 (6.14), 299.1135 (13.15), 223.0603 (117.03) | G 13 (isomer) | [13,30] |
| 36 | 29.28 | C25H28O12 | 519.1508 | 519.1512 | 0.7 | 227.0560 (13.80), 189.0557 (34.00), 183.0664 (34.60), 165.0557 (28.12), 161.0610 (100), 147.0457 (63.08), 121.0669 (46.70) | 6′-O-trans-Cinnamoyl-8-epikingisidic acid | [21] |
| 37 | 30.27 | C48H64O27 | 1071.3562 | 1071.3578 | 1.4 | 909.3159 (15.90), 839.2724 (12.16), 771.2436 (52.95), 685.2404 (100), 523.1850 (63.76), 453.1421 (44.96), 403.1256 (28.24), 385.1157 (15.01), 299.1137 (28.75), 223.0611 (37.36) | G 13 (isomer) | [13,30] |
| 38 | 31.99 | C48H64O27 | 1071.3562 | 1071.3583 | 1.9 | 909.3147 (38.95), 839.2689 (30.69), 771.2422 (23.33), 685.2387 (90.89), 523.1830 (61.19), 453.1407 (42.67), 403.1256 (25.28), 385.1148 (7.88), 399.1131 (22.25), 223.0604 (30.57) | G 13 (isomer) | [13,30] |
| 40 | 34.28 | C25H28O12 | 519.1508 | 519.1510 | 0.3 | 475.1626 (29.18), 209.0447 (14.46), 189.0552 (41.69), 183.0654 (19.17), 165.0565 (29.89), 161.0604 (100), 147.0448 (76.02), 121.0667 (42.59) | 6′-O-cis-Cinnamoyl 8-epikingisidic acid | [21] |
| 41 | 34.54 | C19H22O8 | 377.1242 | 377.1243 | 0.2 | 307.0800 (39.23), 275.0899 (25.15), 149.0273 (100), 139.0394 (77.64) | Oleuropein aglycone | [27,29,32] |
| Triterpenes | ||||||||
| 42 | 39.55 | C30H48O5 | 487.3429 | 487.3424 | −1 | 469.3340 (18.52), 423.3282 (15.84) | Tormentic acid | [33] |
| 43 | 40.97 | C30H48O4 | 471.3480 | 471.3473 | −1.9 | 453.3402 (60.01), 407.3328 (18.05), 451.3225 (13.04) | 19α-Hydroxyursolic acid | [33] |
| 44 | 41.30 | C30H48O4 | 471.3480 | 471.3476 | −0.8 | - | 2α-Hydroxyoleanolic acid | [33] |
| 45 | 41.56 | C30H48O4 | 471.3480 | 471.3475 | −0.9 | - | 2α-Hydroxyursolic acid | [3] |
| 46 | 42.73 | C39H54O6 | 617.3848 | 617.3846 | −0.2 | 145.0292 (14.69) | 3β-O-trans-p-Coumaroylmaslinic acid | [34] |
| 47 | 43.08 | C39H54O6 | 617.3848 | 617.3847 | −0.1 | 145.0288 (17.35) | 3β-O-cis-p-Coumaroylmaslinic acid | [34] |
| 48 | 43.39 | C32H50O5 | 513.3586 | 513.3579 | −1.2 | 495.3497 (27.14), 453.3390 (3.71) | 19α-Hydroxy-3-acetylursolic acid | [34] |
| 49 | 44.35 | C30H48O3 | 455.3531 | 455.3526 | −1 | - | Oleanolic acid/Ursolic acid | standard |
| 50 | 46.71 | C32H50O4 | 497.3636 | 497.3632 | −0.9 | - | Acetyloleanolic acid/Acetylursolic acid | [34] |
| Other Compounds | ||||||||
| 1 | 0.88 | C7H12O6 | 191.0561 | 191.0569 | 4.2 | 173.0447 (11.54), 127.0394 (20.92), 109.0301 (13.44), 93.0359 (67.79), 85.0314 (100) | Quinic acid | [8,19] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Yao, W.; Liu, Q.; Xu, J.; Bao, B.; Shan, M.; Cao, Y.; Cheng, F.; Ding, A.; Zhang, L. Application of UHPLC-ESI-Q-TOF-MS to Identify Multiple Constituents in Processed Products of the Herbal Medicine Ligustri Lucidi Fructus. Molecules 2017, 22, 689. https://doi.org/10.3390/molecules22050689
Li H, Yao W, Liu Q, Xu J, Bao B, Shan M, Cao Y, Cheng F, Ding A, Zhang L. Application of UHPLC-ESI-Q-TOF-MS to Identify Multiple Constituents in Processed Products of the Herbal Medicine Ligustri Lucidi Fructus. Molecules. 2017; 22(5):689. https://doi.org/10.3390/molecules22050689
Chicago/Turabian StyleLi, Hui, Weifeng Yao, Qinan Liu, Jia Xu, Beihua Bao, Mingqiu Shan, Yudan Cao, Fangfang Cheng, Anwei Ding, and Li Zhang. 2017. "Application of UHPLC-ESI-Q-TOF-MS to Identify Multiple Constituents in Processed Products of the Herbal Medicine Ligustri Lucidi Fructus" Molecules 22, no. 5: 689. https://doi.org/10.3390/molecules22050689







