Generation and Applications of a DNA Aptamer against Gremlin-1
Abstract
:1. Introduction
2. Results
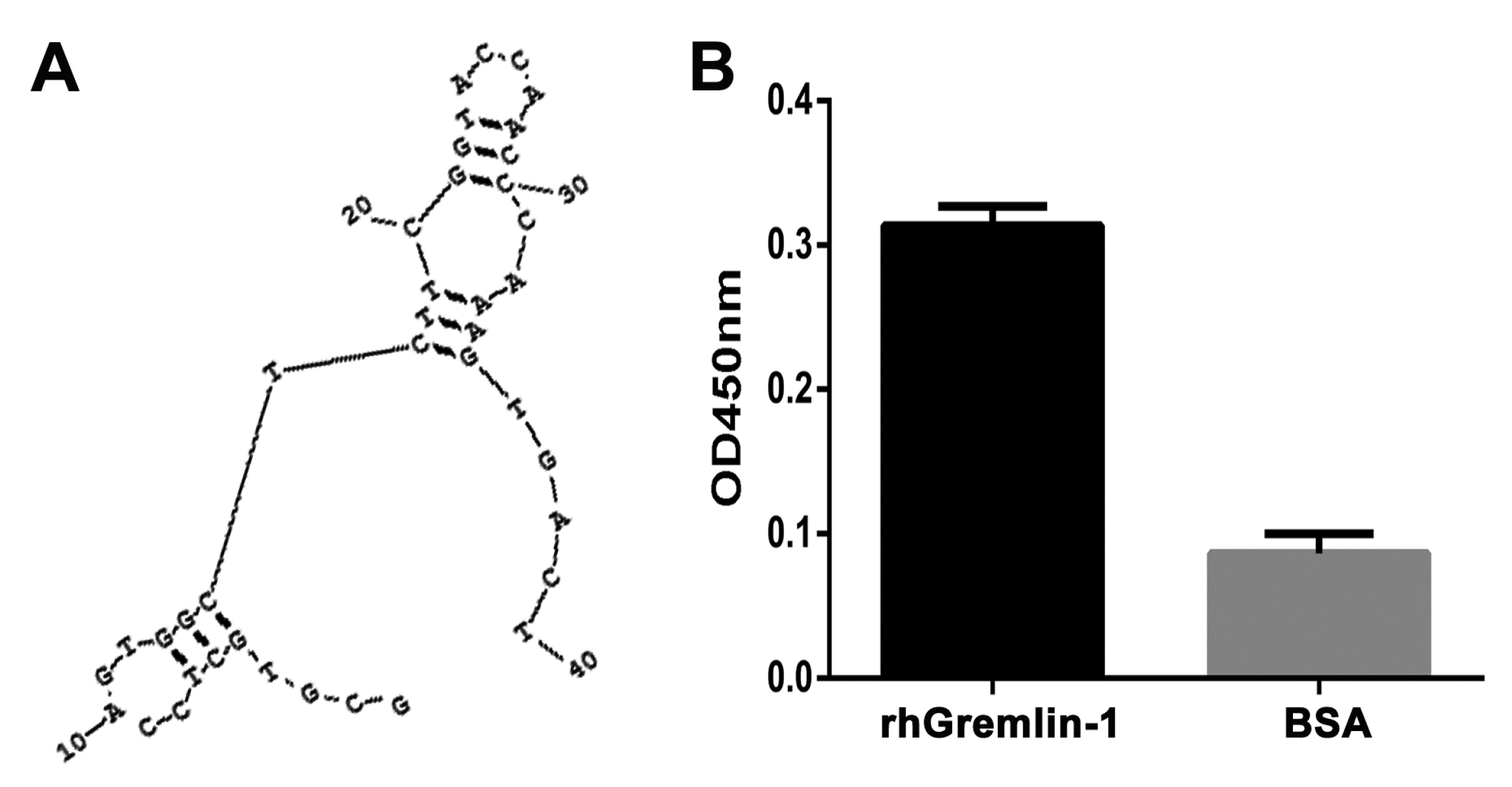
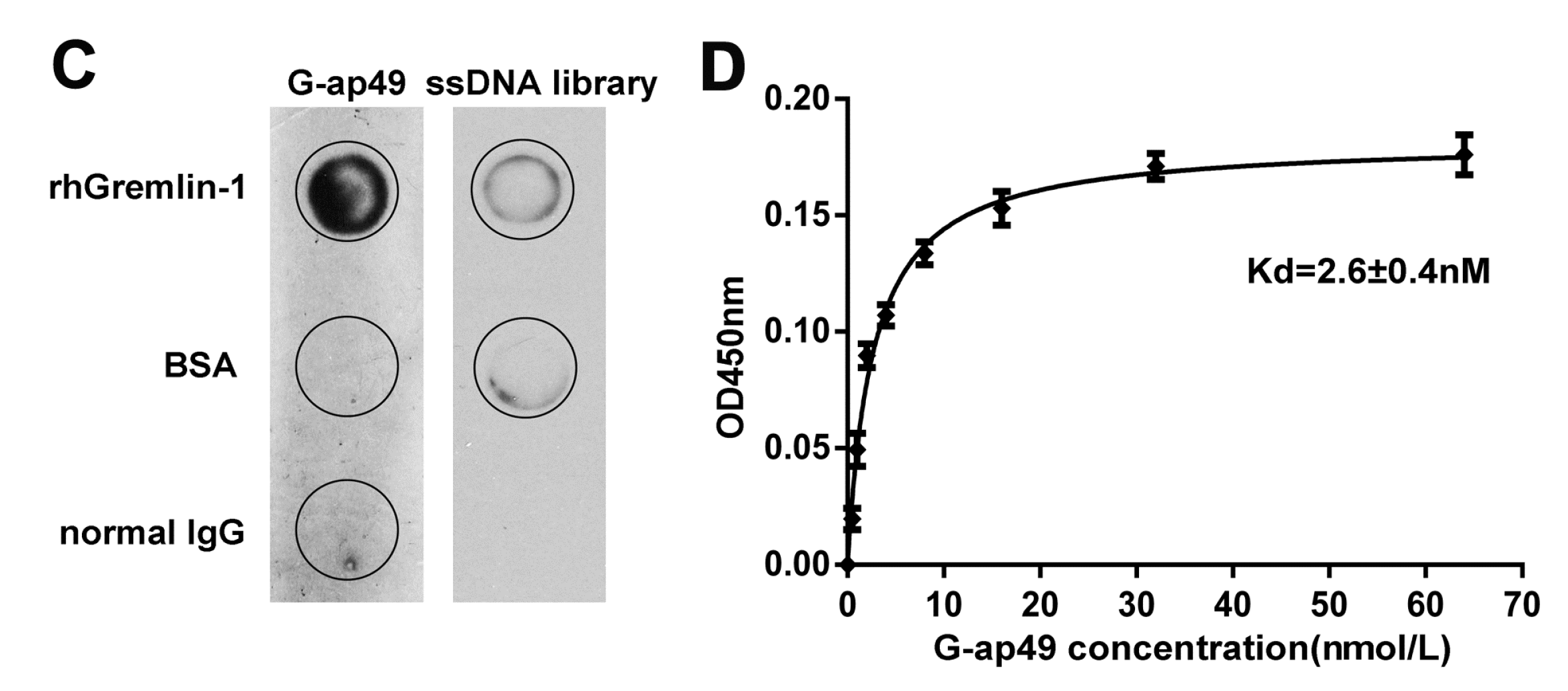
2.1. Selection and Characterization of G-ap49 Specific to Gremlin-1
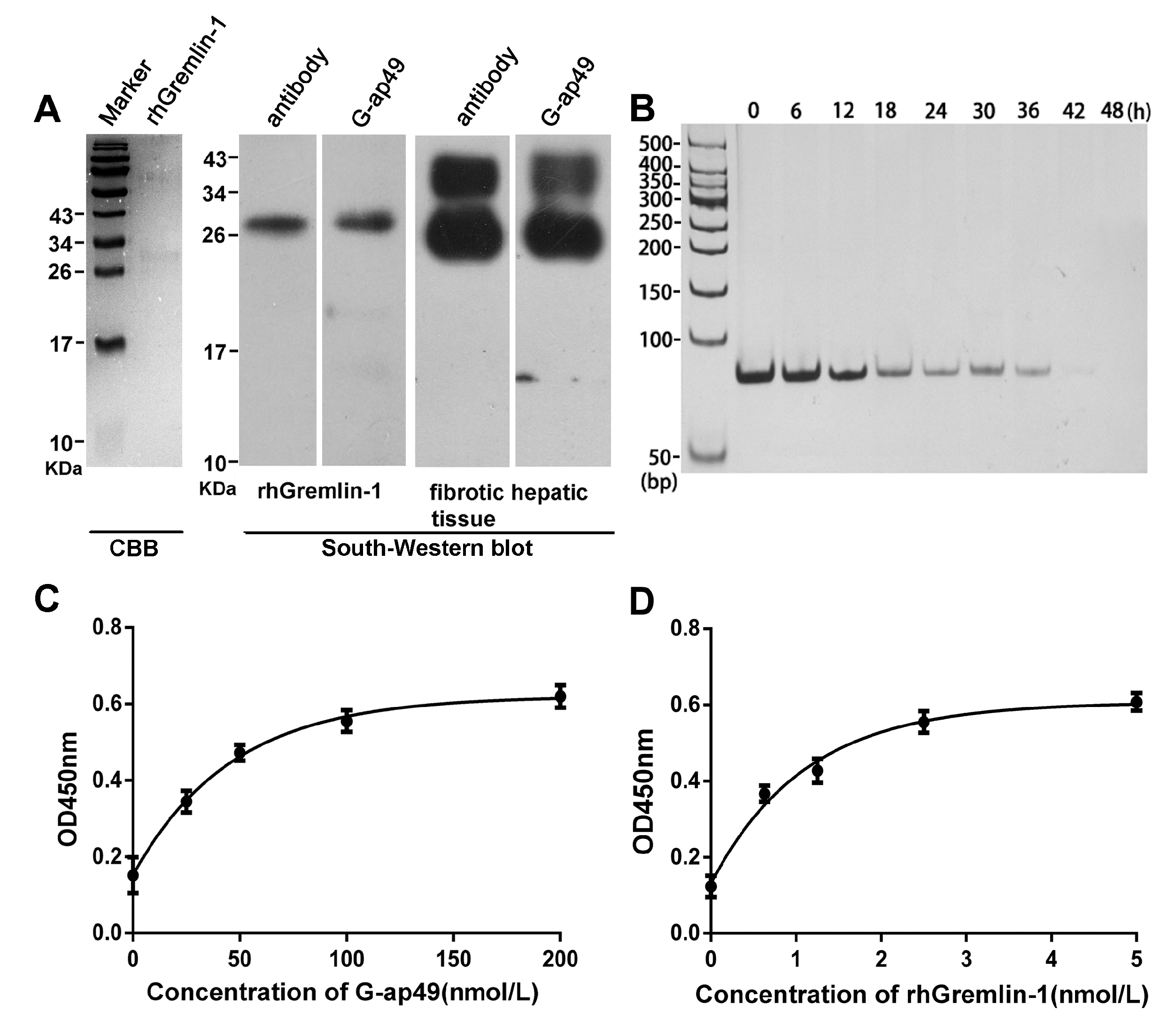
2.2. Both G-ap49 and Anti-Gremlin-1 Antibody Are Effective to Detect Gremlin-1
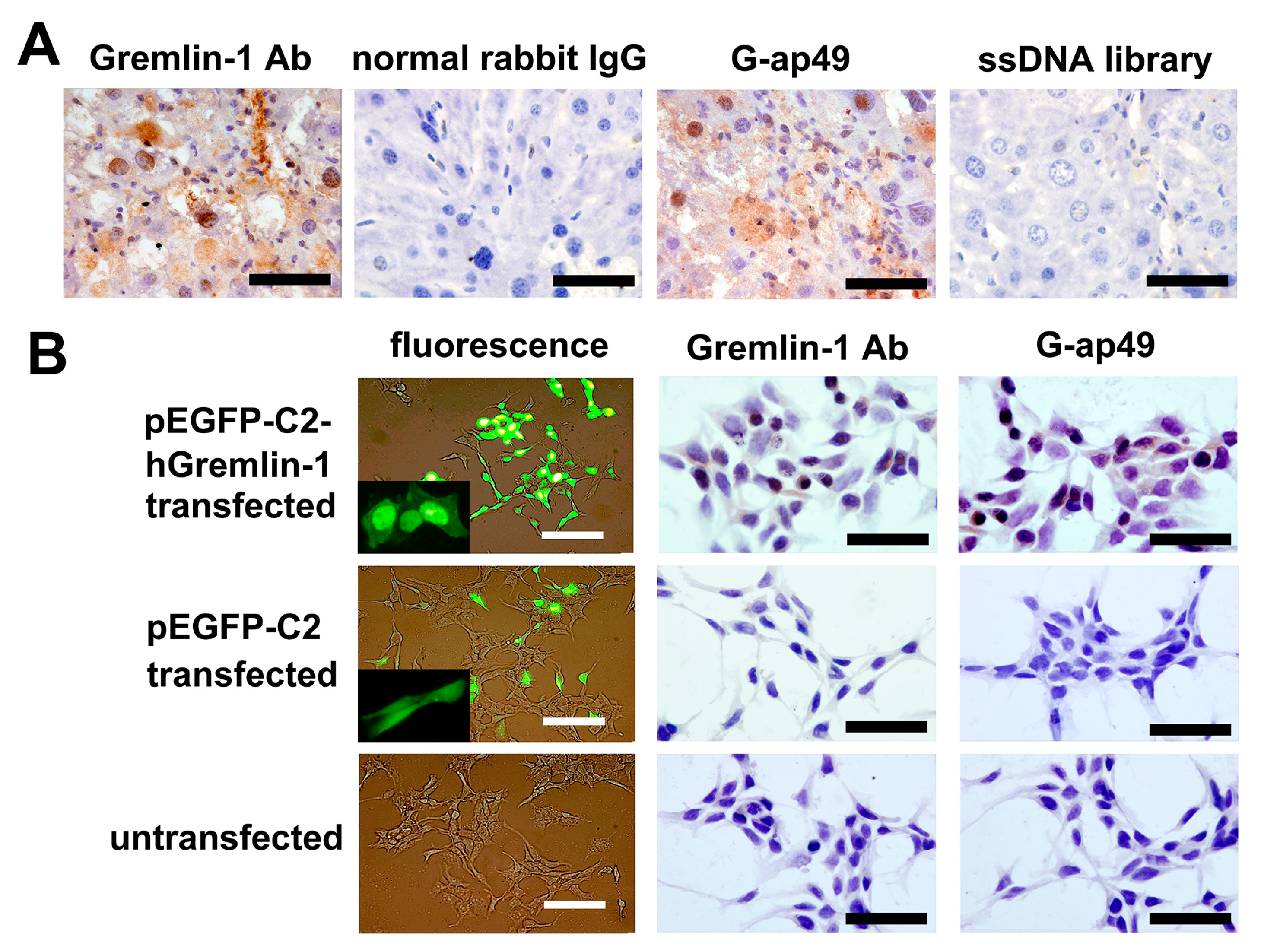
2.3. G-ap49 Was Optimal to Detect Gremlin-1 in Immunohisto-Cytochemical Staining
3. Discussion
4. Materials and Methods
4.1. Initial Library and Primers for SELEX
4.2. SELEX Procedure
4.3. Cloning, Aptamer Binding Assay, DNA Sequencing and Secondary Structure Prediction
4.4. Dot-Blot
4.5. Determination of Equilibrium Dissociation Constants (Kd)
4.6. South-Western Blotting
4.7. ELASA
4.8. Aptamer-Based Histochemistry and Cytochemistry
4.9. Biological Stability Assay
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rider, C.C.; Mulloy, B. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem. J. 2010, 429, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Topol, L.Z.; Bardot, B.; Zhang, Q.; Resau, J.; Huillard, E.; Marx, M.; Calothy, G.; Blair, D.G. Biosynthesis, post-translation modification, and functional characterization of Drm/Gremlin. J. Biol. Chem. 2000, 275, 8785–8793. [Google Scholar] [CrossRef] [PubMed]
- Michos, O.; Panman, L.; Vintersten, K.; Beier, K.; Zeller, R.; Zuniga, A. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development 2004, 131, 3401–3410. [Google Scholar] [CrossRef] [PubMed]
- Stabile, H.; Mitola, S.; Moroni, E.; Belleri, M.; Nicoli, S.; Coltrini, D.; Peri, F.; Pessi, A.; Orsatti, L.; Talamo, F.; et al. Bone morphogenic protein antagonist Drm/Gremlin is a novel proangiogenic factor. Blood 2007, 109, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Chen, S.L.; Lu, X.J.; Shen, C.Y.; Liu, Y.; Chen, Y.P. Bone morphogenetic protein 7 suppresses the progression of hepatic fibrosis and regulates the expression of Gremlin and transforming growth factor β1. Mol. Med. Rep. 2012, 6, 246–252. [Google Scholar] [PubMed]
- Gazzerro, E.; Smerdel-Ramoya, A.; Zanotti, S.; Stadmeyer, L.; Durant, D.; Economides, A.N.; Canalis, E. Conditional deletion of Gremlin causes a transient increase in bone formation and bone mass. J. Biol. Chem. 2007, 282, 31549–31557. [Google Scholar] [CrossRef] [PubMed]
- Costello, C.M.; Cahill, E.; Martin, F.; Gaine, S.; McLoughlin, P. Role of gremlin in the lung: Development and disease. Am. J. Respir. Cell Mol. Biol. 2010, 42, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Claesson-Welsh, L. Gremlin: Vexing VEGF receptor agonist. Blood 2010, 116, 3386–3387. [Google Scholar] [CrossRef] [PubMed]
- Mitola, S.; Ravelli, C.; Moroni, E.; Salvi, V.; Leali, D.; Ballmer-Hofer, K.; Zammataro, L.; Presta, M. Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood 2010, 116, 3677–3680. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, H.; Shin, S.M.; Kim, H.K.; Ha, S.A.; Cho, G.W.; Hur, S.Y.; Kim, T.E.; Kim, J.W. The bone morphogenetic protein antagonist Gremlin-1 is overexpressed in human cancers and interacts with YWHAH protein. BMC Cancer 2006, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Farkas, L.; Farkas, D.; Gauldie, J.; Warburton, D.; Shi, W.; Kolb, M. Transient overexpression of Gremlin results in epithelial activation and reversible fibrosis in rat lungs. Am. J. Respir. Cell Mol. Biol. 2011, 44, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Myllärniemi, M.; Lindholm, P.; Ryynänen, M.J.; Kliment, C.R.; Salmenkivi, K.; Keski-Oja, J.; Kinnula, V.L.; Oury, T.D.; Koli, K. Gremlin-mediated decrease in bone morphogenetic protein signaling promotes pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2008, 177, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.; McMahon, R.; Lappin, D.W.; Brady, H.R. Gremlins: Is this what renal fibrogenesis has come to? Exp. Nephrol. 2002, 10, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Droguett, A.; Krall, P.; Burgos, M.E.; Valderrama, G.; Carpio, D.; Ardiles, L.; Rodriguez-Diez, R.; Kerr, B.; Walz, K.; Ruiz-Ortega, M.; et al. Tubular overexpression of Gremlin induces renal damage susceptibility in mice. PLoS ONE 2014, 9, e101879. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, G.; Droguett, A.; Burgos, M.E.; Aros, C.; Ardiles, L.; Flores, C.; Carpio, D.; Ruiz-Ortega, M.; Egido, J.; Mezzano, S. Gremlin: A novel mediator of epithelial mesenchymal transition and fibrosis in chronic allograft nephropathy. Transpl. Proc. 2008, 40, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Sneddon, J.B.; Zhen, H.H.; Montgomery, K.; van de Rijn, M.; Tward, A.D.; West, R.; Gladstone, H.; Chang, H.Y.; Morganroth, G.S.; Oro, A.E.; et al. Bone morphogenetic protein antagonist Gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proc. Natl. Acad. Sci. USA 2006, 103, 14842–14847. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, E.; McNamara, J., II; Pastor, F. Use of oligonucleotide aptamer ligands to modulate the function of immune receptors. Clin. Cancer Res. 2013, 19, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.K.; Cheng, M.L.; Wu, R.M.; Yao, Y.M.; Mu, M.; Zhu, J.J.; Zhang, B.F.; Zhou, M.Y. Effect of Danshao Huaxian capsule on Gremlin and bone morphogenetic protein-7 expression in hepatic fibrosis in rats. World J. Gastroenterol. 2014, 20, 14875–14883. [Google Scholar] [CrossRef] [PubMed]
- Wellbrock, J.; Sheikhzadeh, S.; Oliveira-Ferrer, L.; Stamm, H.; Hillebrand, M.; Keyser, B.; Klokow, M.; Vohwinkel, G.; Bonk, V.; Otto, B.; et al. Overexpression of Gremlin-1 in patients with Loeys-Dietz syndrome: Implications on pathophysiology and early disease detection. PLoS ONE 2014, 9, e104742. [Google Scholar] [CrossRef] [PubMed]
- Han, E.J.; Yoo, S.A.; Kim, G.M.; Hwang, D.; Cho, C.S.; You, S.; Kim, W.U. GREM1 is a key regulator of synoviocyte hyperplasia and invasiveness. J. Rheumatol. 2016, 43, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.A.; Rath, D.; Schmid, M.; Schoenleber, H.; Gawaz, M.; Geisler, T.; Müller, I.I. High plasma levels of Gremlin-1 and macrophage migration inhibitory factor, but not their ratio, indicate an increased risk for acute coronary syndrome in patients with type 2 diabetes mellitus. Clin. Cardiol. 2016, 39, 201–206. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compound recombinant plasmid pEGFP-C2-hGremlin-1 are available from the authors. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Huo, Y.; Guo, Y.; Zheng, X.; Sun, W.; Hao, Z. Generation and Applications of a DNA Aptamer against Gremlin-1. Molecules 2017, 22, 706. https://doi.org/10.3390/molecules22050706
Li Q, Huo Y, Guo Y, Zheng X, Sun W, Hao Z. Generation and Applications of a DNA Aptamer against Gremlin-1. Molecules. 2017; 22(5):706. https://doi.org/10.3390/molecules22050706
Chicago/Turabian StyleLi, Qian, Yongwei Huo, Yonghong Guo, Xiaoyan Zheng, Wengang Sun, and Zhiming Hao. 2017. "Generation and Applications of a DNA Aptamer against Gremlin-1" Molecules 22, no. 5: 706. https://doi.org/10.3390/molecules22050706





