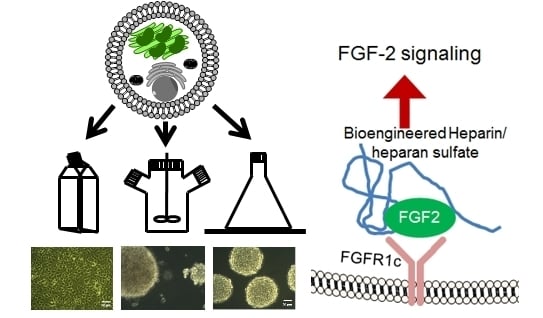
Structure-Activity Relationships of Bioengineered Heparin/Heparan Sulfates Produced in Different Bioreactors
Abstract
:1. Introduction
2. Results
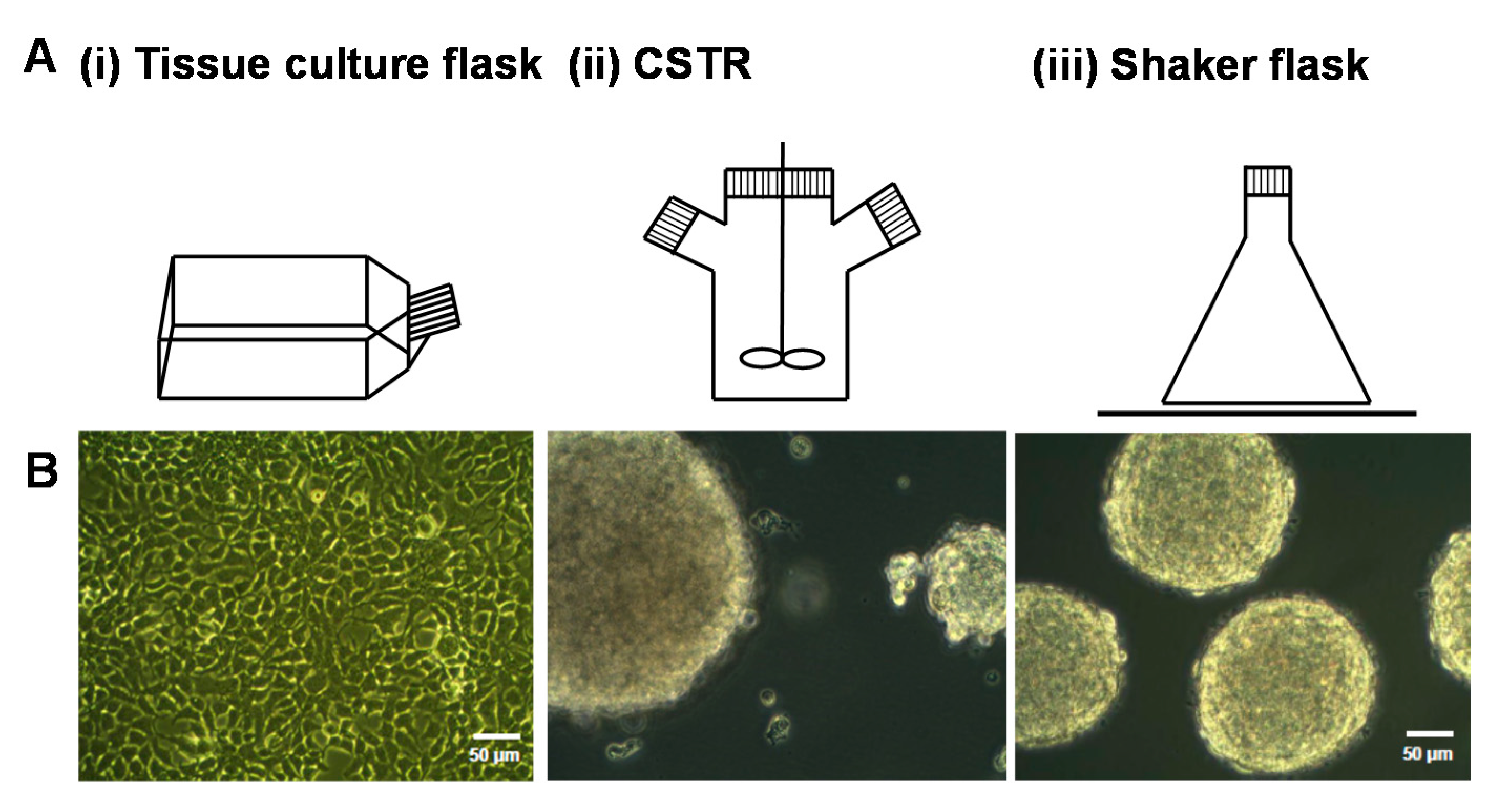
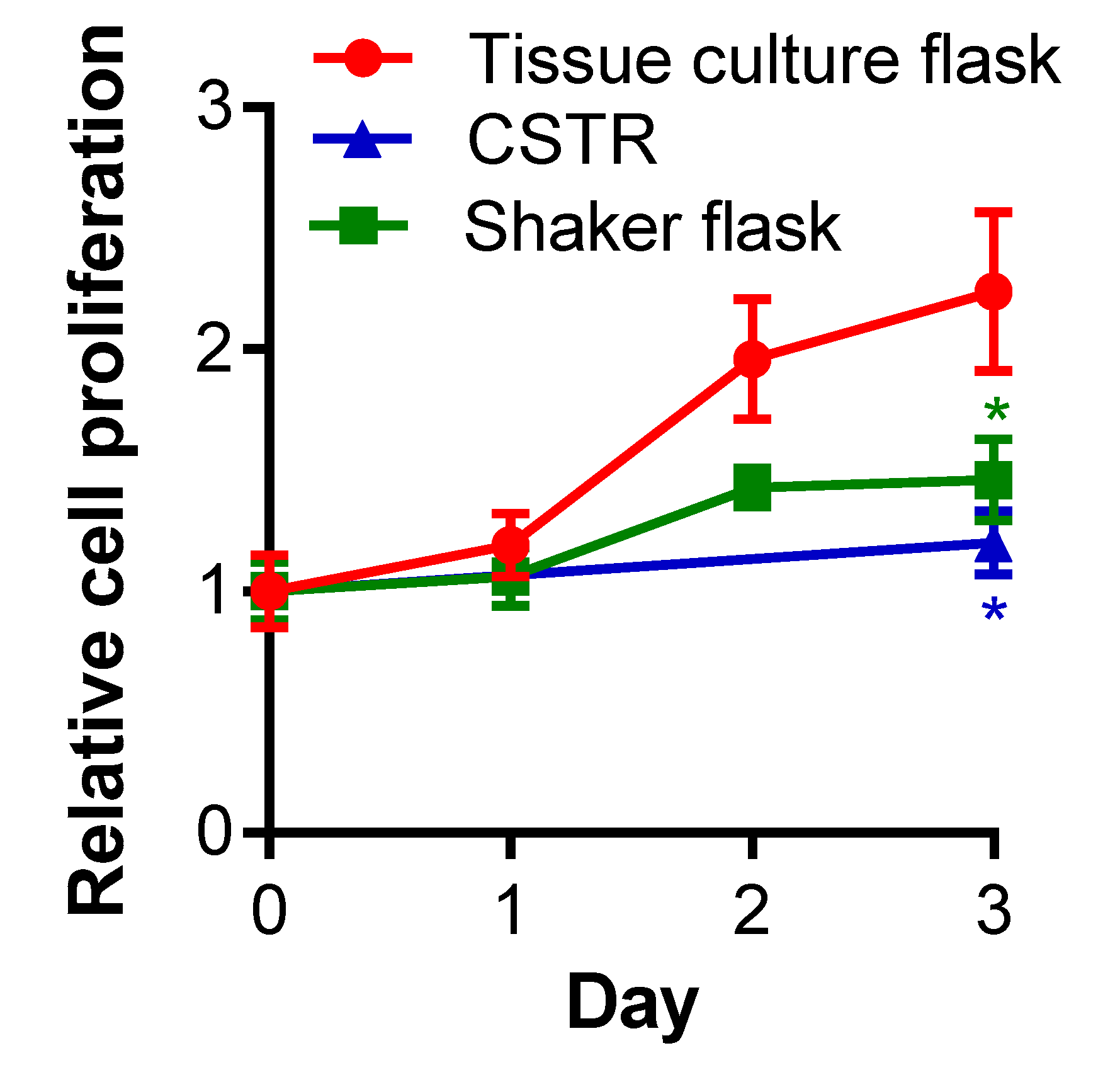
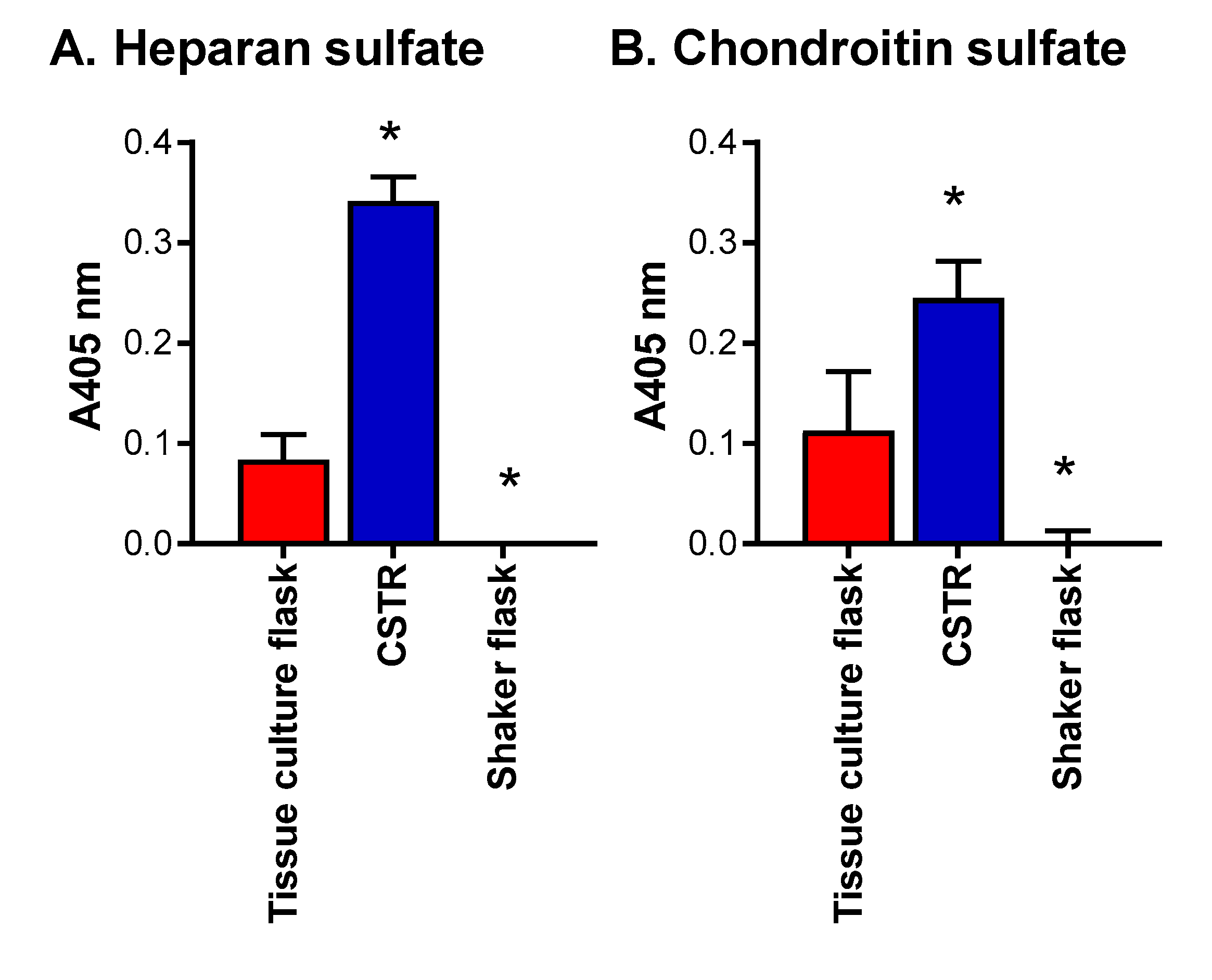
2.1. The Effect of Different Bioreactors on Serglycin Production
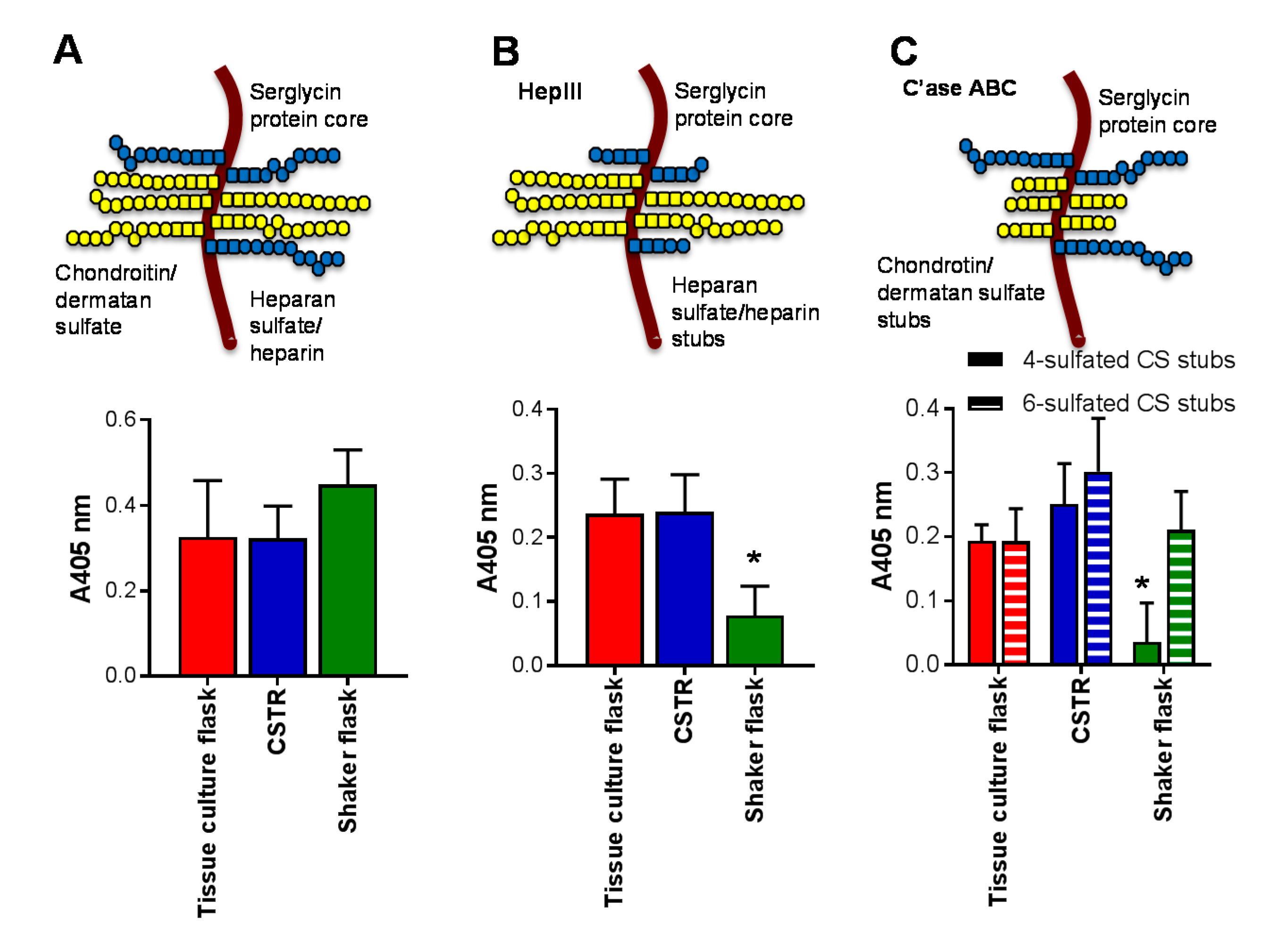
2.2. The Effect of Different Culture Conditions on Serglycin Production
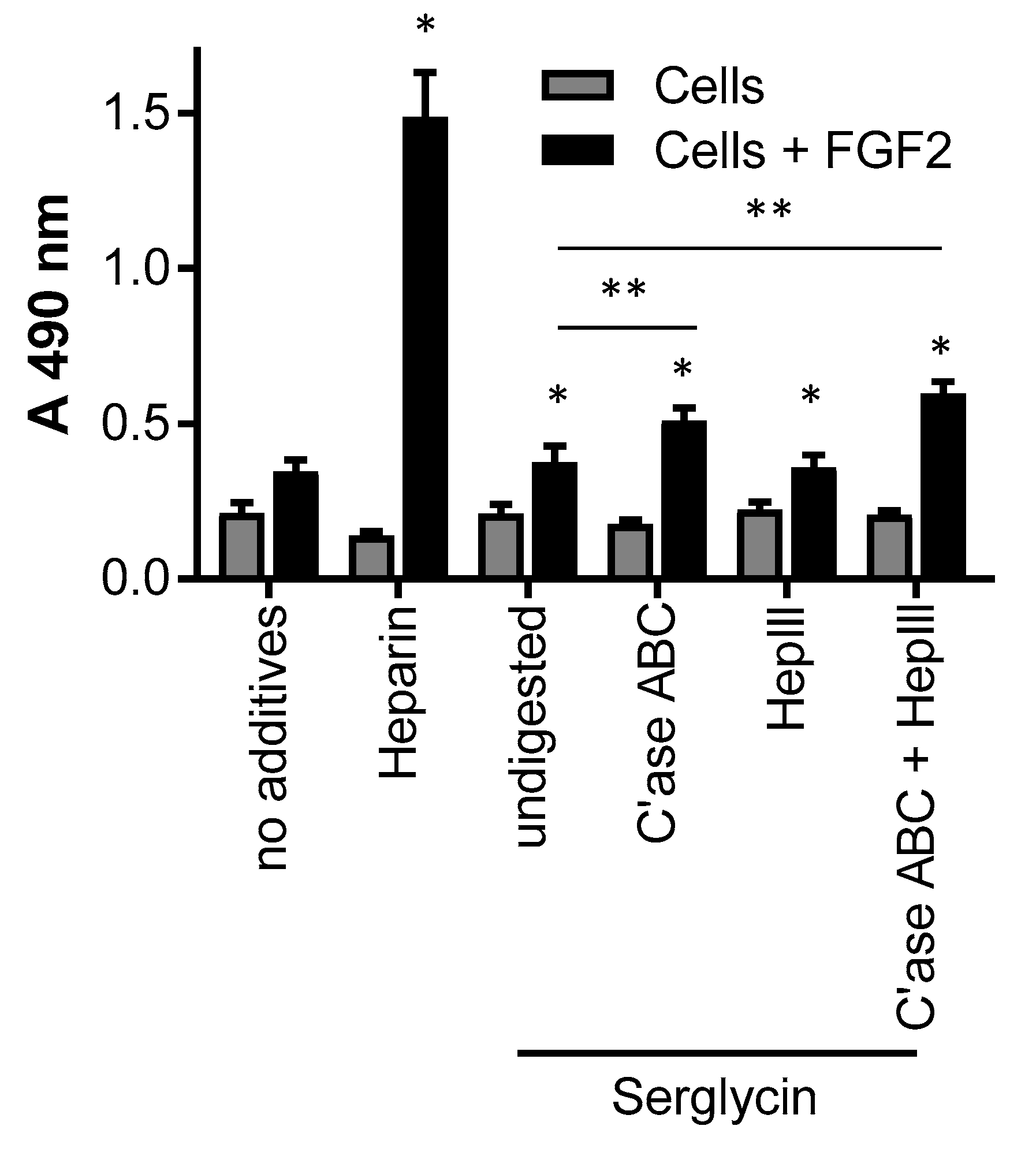
2.3. The Effect of Serglycin Glycosaminoglycan Decoration on Growth Factor Binding and Signaling
3. Discussion
4. Materials and Methods
4.1. Culture of Mammalian Cells Expressing Serglycin
4.2. Cell Proliferation Assay
4.3. Isolation of Serglycin
4.4. Glycosaminoglycan Digestion
4.5. ELISA
4.6. Growth Factor Binding and Signaling of FGF-2
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lord, M.S.; Cheng, B.; Tang, F.; Lyons, J.G.; Rnjak-Kovacina, J.; Whitelock, J.M. Bioengineered human heparin with anticoagulant activity. Metab. Eng. 2016, 38, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, U.; Backstrom, G.; Hook, M.; Thunberg, L.; Fransson, L.A.; Linker, A. Structure of the antithrombin-binding site in heparin. Proc. Natl. Acad. Sci. USA 1979, 76, 3198–3202. [Google Scholar] [CrossRef] [PubMed]
- Linhardt, R.J. 2003 Claude S. Hudson Award address in carbohydrate chemistry. Heparin: Structure and activity. J. Med. Chem. 2003, 46, 2551–2564. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, U.; Sterner, E.; Hickey, A.M.; Onishi, A.; Zhang, F.; Dordick, J.S.; Linhardt, R.J. Engineering of routes to heparin and related polysaccharides. Appl. Microbiol. Biotechnol. 2012, 93, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sakiyama-Elbert, S.E. Incorporation of heparin into biomaterials. Acta Biomater. 2014, 10, 1581–1587. [Google Scholar] [CrossRef] [PubMed]
- Lever, R.; Page, C.P. Novel drug development opportunities for heparin. Nat. Rev. Drug Discov. 2002, 1, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Toida, T.; Yoshida, H.; Toyoda, H.; Koshiishi, I.; Imanari, T.; Hileman, R.E.; Fromm, J.R.; Linhardt, R.J. Structural differences and the presence of unsubstituted amino groups in heparan sulphates from different tissues and species. Biochem. J. 1997, 322, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Kolset, S.O.; Gallagher, J. Proteoglycans in haemopoietic cells. Biochim. Biophys. Acta 1990, 1032, 191–221. [Google Scholar] [CrossRef]
- Kolset, S.O.; Mann, D.M.; Uhlin-Hansen, L.; Winberg, J.O.; Ruoslahti, E. Serglycin-binding proteins in activated macrophages and platelets. J. Leukoc. Biol. 1996, 59, 545–554. [Google Scholar] [PubMed]
- Kolset, S.O.; Pejler, G. Serglycin: A structural and functional chameleon with wide impact on immune cells. J. Immunol. 2011, 187, 4927–4933. [Google Scholar] [CrossRef] [PubMed]
- Kolset, S.O.; Tveit, H. Serglycin—Structure and biology. Cell. Mol. Life Sci. 2008, 65, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Wang, H.; Bernfield, M.; Gallagher, J.T.; Turnbull, J.E. Cell surface syndecan-1 on distinct cell types differs in fine structure and ligand binding of its heparan sulfate chains. J. Biol. Chem. 1994, 269, 18881–18890. [Google Scholar] [PubMed]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Bernfield, M.; Gotte, M.; Park, P.W.; Reizes, O.; Fitzgerald, M.L.; Lincecum, J.; Zako, M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–777. [Google Scholar] [CrossRef] [PubMed]
- Lin, X. Fuctions of heparan sulfate proteoglycans in cell signaling during development. Development 2004, 131, 6009–6021. [Google Scholar] [CrossRef] [PubMed]
- Salmivirta, M.; Lidholt, K.; Linhadl, U. Heparan sulfate: A piece of information. FASEB J. 1996, 10, 1270–1279. [Google Scholar] [PubMed]
- Turnbull, J.E. Heparan sulfate glycomics: Towards systems biology strategies. Biochem. Soc. Trans. 2010, 38, 1356–1360. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, R.D.; Shworak, N.W.; Liu, J.; Schwartz, J.J.; Zhang, L. Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? J. Clin. Investig. 1997, 99, 2062–2070. [Google Scholar] [CrossRef] [PubMed]
- Kreuger, J.; Salmivirta, M.; Sturiale, L.; Gimenez-Gallego, G.; Lindahl, U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J. Biol. Chem. 2001, 276, 30744–30752. [Google Scholar] [CrossRef] [PubMed]
- Linhardt, R.J.; Gunay, N.S. Production and chemical processing of low molecular weight heparins. Semin. Thromb. Hemost. 1999, 25, 5–16. [Google Scholar] [PubMed]
- Ashikari-Hada, S.; Habuchi, H.; Kariya, Y.; Itoh, N.; Reddi, A.H.; Kimata, K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J. Biol. Chem. 2004, 279, 12346–12354. [Google Scholar] [CrossRef] [PubMed]
- Yates, E.A.; Guimond, S.E.; Turnbull, J.E. Highly diverse heparan sulfate analogue libraries: Providing access to expanded areas of sequence space for bioactivity screening. J. Med. Chem. 2004, 47, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.K.; Ahmed, Y.A.; Yates, E.A.; Turnbull, J.E. Generating heparan sulfate saccharide libraries for glycomics applications. Nat. Protoc. 2010, 5, 821–833. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, P.L.; Liu, J.; Linhardt, R.J. Chemoenzymatic synthesis of glycosaminoglycans: Re-creating, re-modeling and re-designing nature's longest or most complex carbohydrate chains. Glycobiology 2013, 23, 764–777. [Google Scholar] [CrossRef] [PubMed]
- Choay, J.; Petitou, M.; Lormeau, J.C.; Sinaÿ, P.; Casu, B.; Gatti, G. Structure-activity relationship in heparin: A synthetic pentasaccharide with high affinity for antithrombin III and eliciting high anti-factor Xa activity. Biochem. Biophys. Res. Commun. 1983, 116, 492–499. [Google Scholar] [CrossRef]
- Farrugia, B.L.; Lord, M.S.; Melrose, J.; Whitelock, J.M. Can we produce heparin/heparan sulfate biomimetics using ‘mother-nature‘ as the gold standard? Molecules 2015, 20, 4254–4276. [Google Scholar] [CrossRef] [PubMed]
- Baik, J.Y.; Gasimli, L.; Yang, B.; Datta, P.; Zhang, F.; Glass, C.A.; Esko, J.D.; Linhardt, R.J.; Sharfstein, S.T. Metabolic engineering of Chinese hamster ovary cells: Towards a bioengineered heparin. Metab. Eng. 2012, 14, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Lord, M.S.; Cheng, B.; Farrugia, B.L.; McCarthy, S.; Whitelock, J.M. Platelet factor 4 binds to vascular proteoglycans and controls both growth factor activities and platelet activation. J. Biol. Chem. 2017, 292, 4054–4063. [Google Scholar] [CrossRef] [PubMed]
- Eibl, R.; Kaiser, S.; Lombriser, R.; Eibl, D. Disposble bioreactors: The current state-of-the-art and recommended applications in biotechnology. Appl. Microbiol. Biotechnol. 2010, 86, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Meuwly, F.; Ruffieux, P.A.; Kadouri, A.; von Stockar, U. Packed-bed bioreactors for mammalian cell culture: Bioprocess and biomedical applications. Biotechnol. Adv. 2007, 25, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Lord, M.S.; Jung, M.; Whitelock, J.M. Optimization of bioengineered heparin/heparan sulfate production for therapeutic applications. Bioengineered 2017, 10, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, T. Effects of glucose on the production of recombinant protein C in mammalian cell culture. Biotechnol. Bioeng. 1992, 39, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Whitford, W.G. Fed-batch mammalian cell culture in bioproduction. BioProcess. Int. 2006, 4, 30–40. [Google Scholar]
- Cechowska-Pasko, M.; Bańkowski, E. Glucose deficiency inhibits glycosaminoglycans synthesis in fibroblast cultures. Biochimie 2010, 92, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Midura, R.J.; Vasanji, A.; Wang, A.J.; Hascall, V.C. Hyperglycemia diverts dividing osteoblastic precursor cells to an adipogenic pathway and induces synthesis of a hyaluronan matrix that is adhesive for monocytes. J. Biol. Chem. 2014, 289, 11410–11420. [Google Scholar] [CrossRef] [PubMed]
- Vogl-Willis, C.A.; Edwards, I.J. High-glucose-induced structural changes in the heparan sulfate proteoglycan, perlecan, of cultured human aortic endothelial cells. Biochim. Biophys. Acta 2004, 1672, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.T. Heparan sulfate: Growth control with a restricted sequence menu. J. Clin. Investig. 2001, 108, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Lyon, M.; Gallagher, J.T. Distinct substrate specificities of bacterial heparinases against N-unsubstituted glucosamine residues in heparan sulfate. J. Biol. Chem. 2005, 280, 15742–15748. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.Y.; Lord, M.S.; Melrose, J.; Rees, M.D.; Knox, S.M.; Freeman, C.; Iozzo, R.V.; Whitelock, J.M. Heparan sulfate dependent signaling of fibroblast growth factor (FGF) 18 by chondrocyte-derived perlecan. Biochemistry 2010, 49, 5524–5532. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.L.; West, L.A.; Govindraj, P.; Zhang, X.; Ornitz, D.M.; Hassell, J.R. Heparan and chondroitin sulfate on growth plate perlecan mediate binding and delivery of FGF-2 to FGF receptors. Matrix Biol. 2007, 26, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.P.; Roubin, R.H.; Whitelock, J.M. Characterization and purification of glycosaminoglycans from crude biological samples. J. Agric. Food Chem. 2008, 56, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Yayon, A.; Flanagan, J.G.; Svahn, C.M.; Levi, E.; Leder, P. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol. Cell Biol. 1992, 12, 240–247. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available upon request from the authors. |


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.N.; Whitelock, J.M.; Lord, M.S. Structure-Activity Relationships of Bioengineered Heparin/Heparan Sulfates Produced in Different Bioreactors. Molecules 2017, 22, 806. https://doi.org/10.3390/molecules22050806
Kim HN, Whitelock JM, Lord MS. Structure-Activity Relationships of Bioengineered Heparin/Heparan Sulfates Produced in Different Bioreactors. Molecules. 2017; 22(5):806. https://doi.org/10.3390/molecules22050806
Chicago/Turabian StyleKim, Ha Na, John M. Whitelock, and Megan S. Lord. 2017. "Structure-Activity Relationships of Bioengineered Heparin/Heparan Sulfates Produced in Different Bioreactors" Molecules 22, no. 5: 806. https://doi.org/10.3390/molecules22050806