An All-Nanocrystal Biosensing System for In Vitro Detection of STAT3 Oligonucleotides
Abstract
:1. Introduction
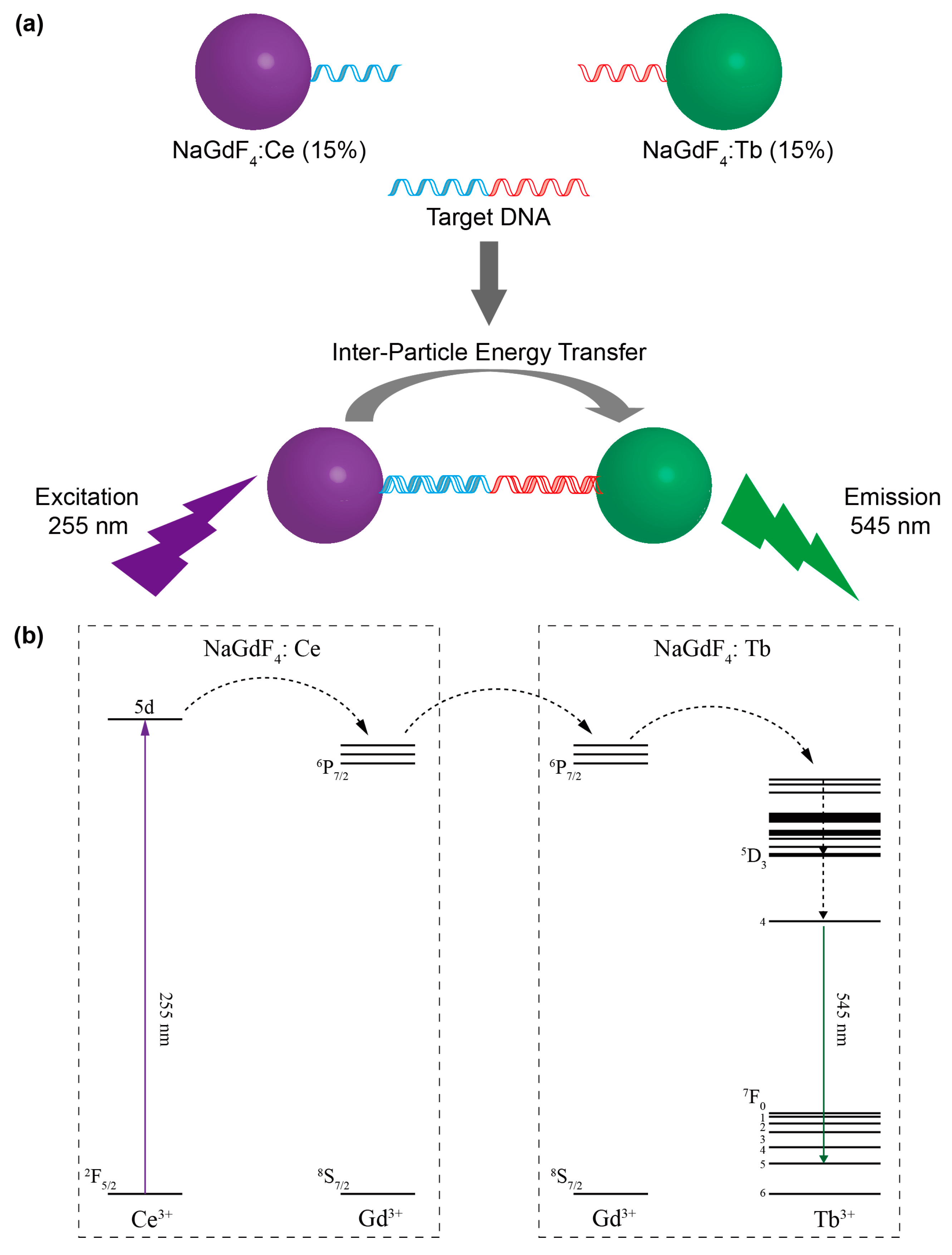
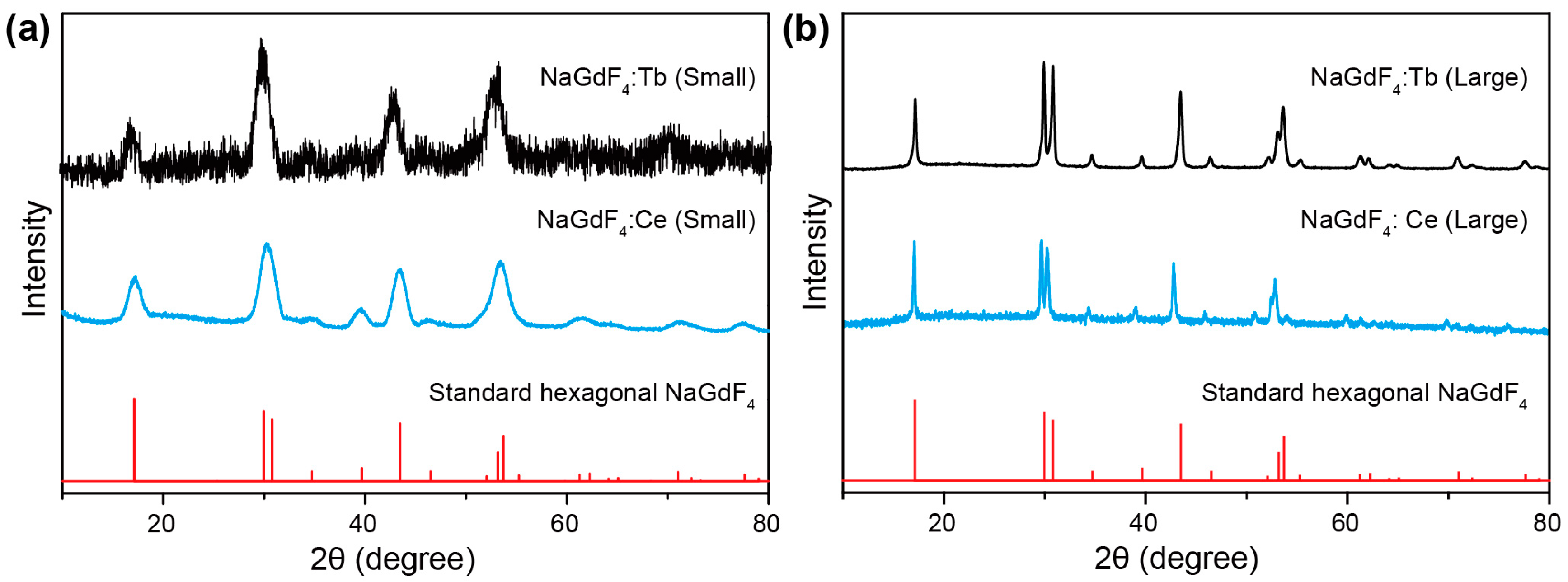
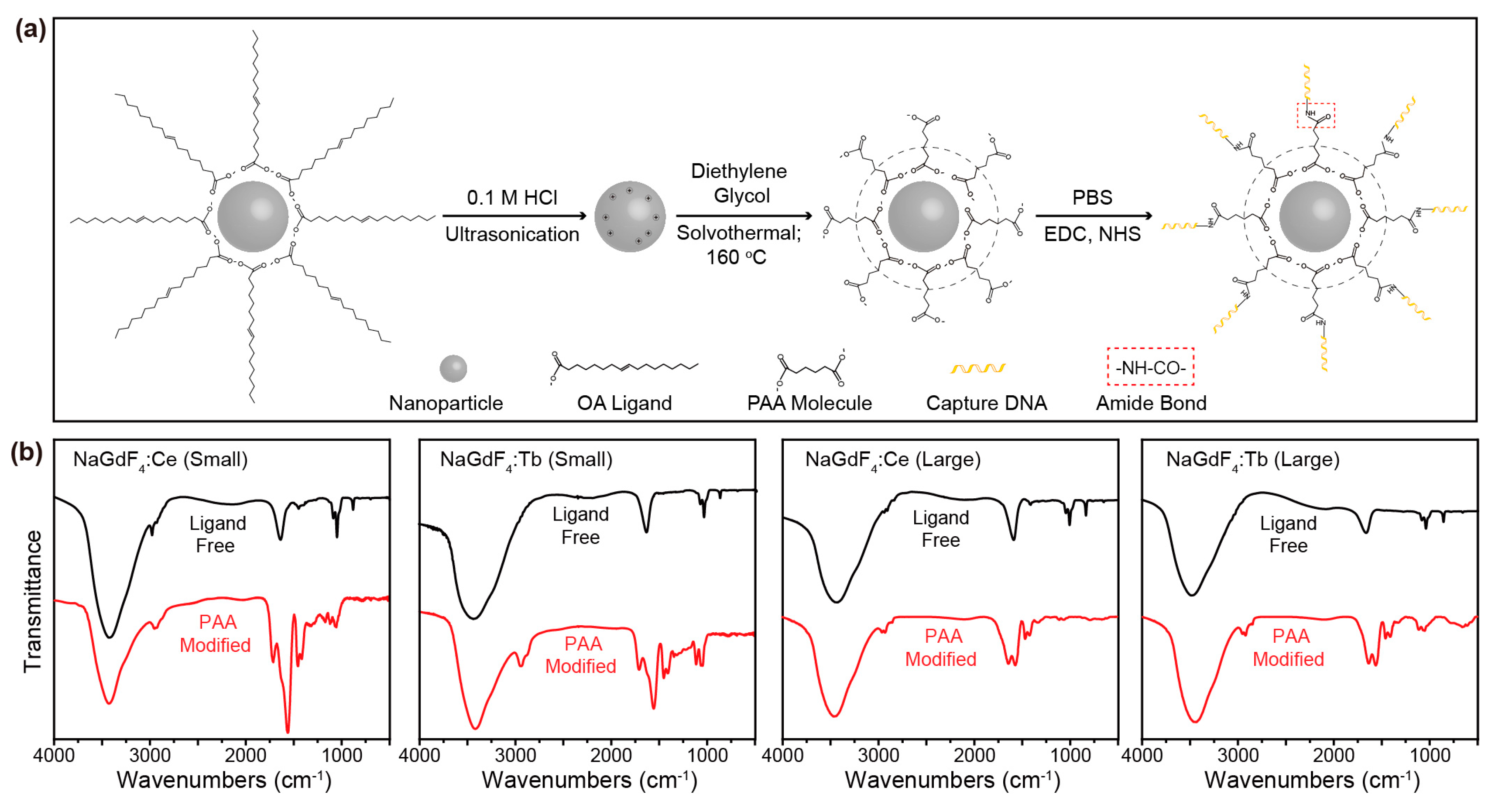
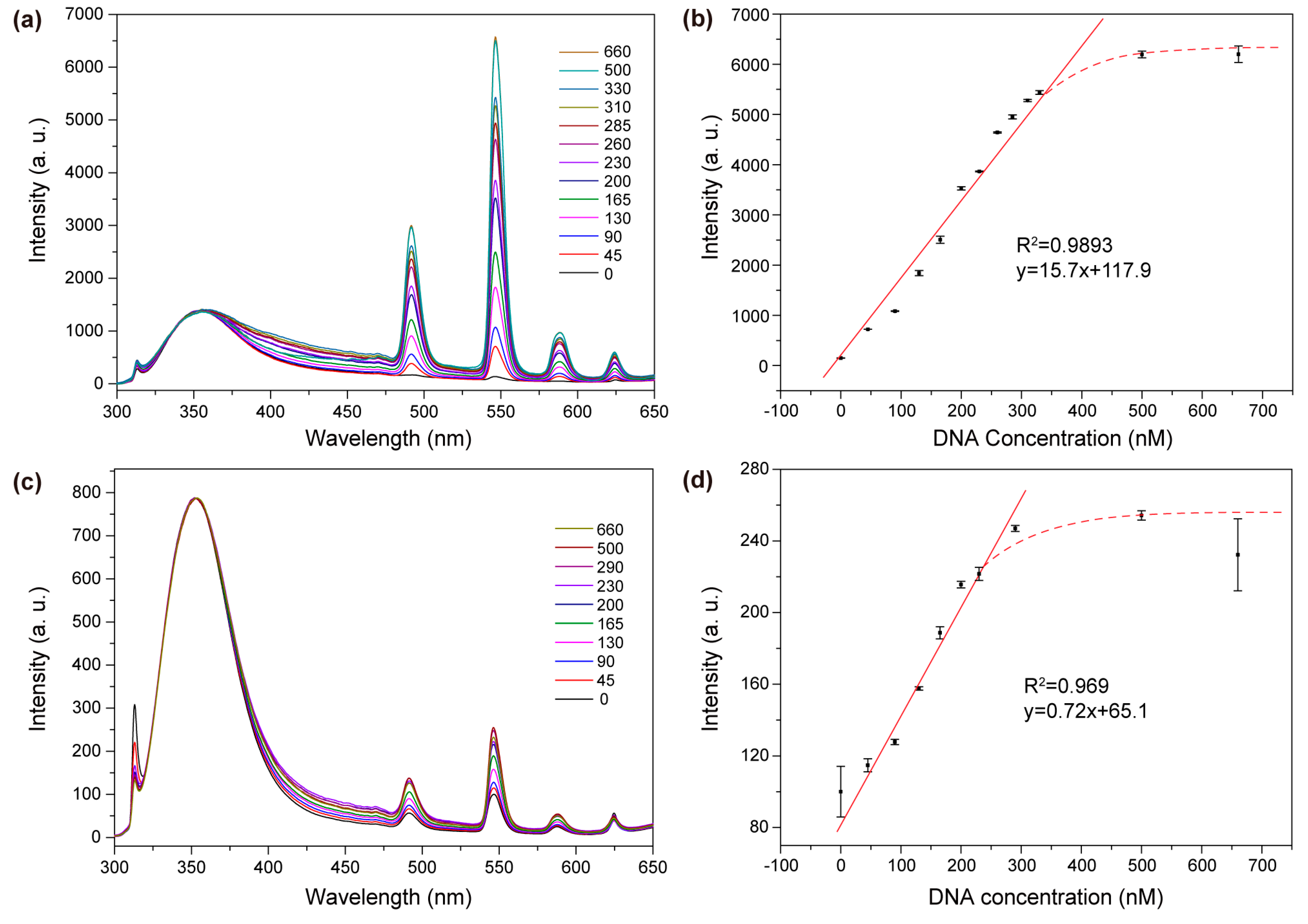
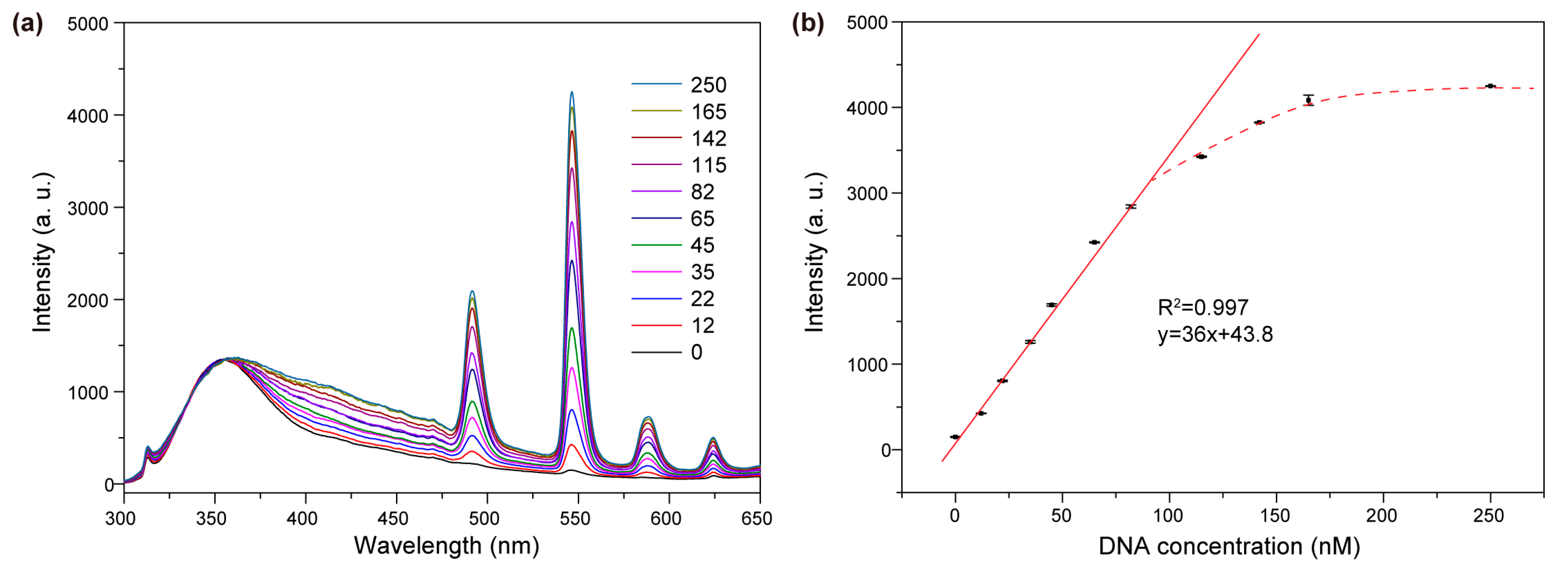
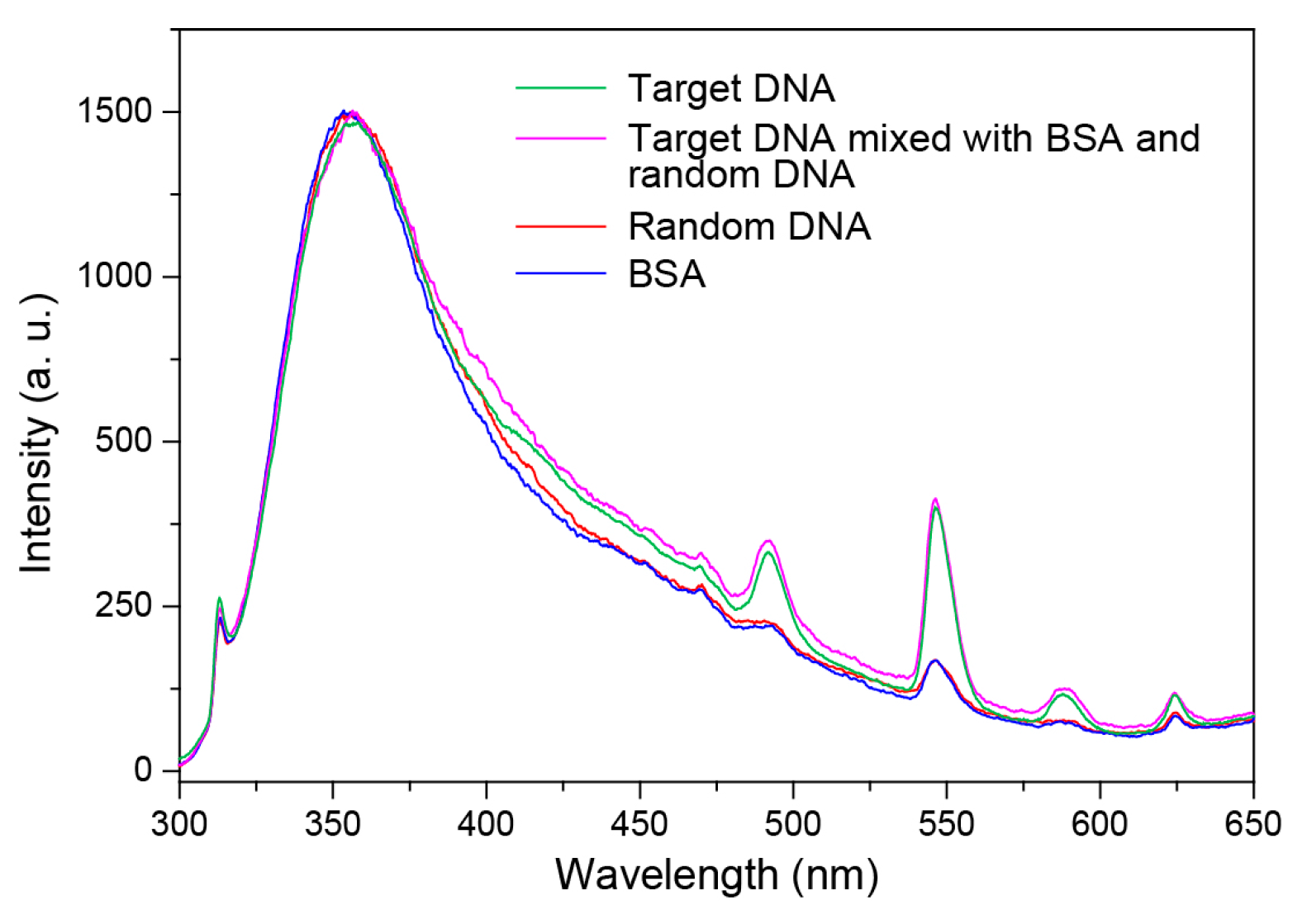
2. Results and Discussion
3. Materials and Methods
3.1. Reagents
3.2. Synthesis of Small NaGdF4:Ln (15%) (Ln = Ce and Tb) Nanocrystals
3.3. Synthesis of Large NaGdF4:Ln (15%) (Ln = Ce and Tb) Nanocrystals
3.4. Synthesis of Ligand-Free Nanocrystals
3.5. Surface Modification of Nanocrystals with PAA
3.6. Synthesis of Capture DNA-Conjugated Nanocrystals
3.7. Detection of STAT3 by In Vitro Assay
3.8. Characterizations
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, G.; Shen, J.; Ohulchanskyy, T.Y.; Patel, N.J.; Kutikov, A.; Li, Z.; Song, J.; Pandey, R.K.; Ågren, H.; Prasad, P.N.; et al. (α-NaYbF4:Tm3+)/CaF2 Core/Shell Nanoparticles with Efficient Near-Infrared to Near-Infrared Upconversion for High-Contrast Deep Tissue Bioimaging. ACS Nano 2012, 6, 8280–8287. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.I.; Lee, K.T.; Suh, Y.D.; Hyeon, T. Upconverting Nanoparticles: A Versatile Platform for Wide-Dield Two-Photon Microscopy and Multi-Modal in Vivo Imaging. Chem. Soc. Rev. 2015, 44, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jin, L.; Kong, W.; Sun, T.; Zhang, W.; Liu, X.; Fan, J.; Yu, S.F.; Wang, F. Confining Energy Migration in Upconversion Nanoparticles towards Deep Ultraviolet Lasing. Nat. Commun. 2016, 7, 10304–10310. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.M.; Chen, X.; Chun, K.S.; Wang, F.; Yu, S.F. Enhancing Multiphoton Upconversion from NaYF4:Yb/Tm@NaYF4 Core-Shell Nanoparticles via the Use of Laser Cavity. ACS Nano 2017, 11, 843–849. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Pang, X.; Liu, X.; Jiang, B.; He, Y.; Snaith, H.; Lin, Z. Monodisperse Dual-Functional Upconversion Nanoparticles enabled Near-Infrared Organolead Halide Perovskite Solar Cells. Angew. Chem. Int. Ed. 2016, 55, 4280–4284. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Ning, Y.; Wu, S.; Niu, W.; Zhang, S. Effectively Utilizing NIR Light Using Direct Electron Injection from Up-Conversion Nanoparticles to the TiO2 Photoanode in Dye-Sensitized Solar Cells. Adv. Funct. Mater. 2013, 23, 5910–5915. [Google Scholar] [CrossRef]
- Oppong, S.O.B.; Anku, W.W.; Shukla, S.K.; Govender, P.P. Lanthanum doped-TiO2 decorated on graphene oxide nanocomposite: A photocatalyst for enhanced degradation of acid blue 40 under simulated solar lightAdvanced. Adv. Mater. Lett. 2017, 8, 295–302. [Google Scholar] [CrossRef]
- Ong, L.C.; Ang, L.Y.; Alonso, S.; Zhang, Y. Bacterial Imaging with Photostable Upconversion Fluorescent Nanoparticles. Biomaterials 2014, 35, 2987–2998. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.-K.; Ye, W.; Wang, G.; Li, J.; Yang, M.; Hao, J. Ultrasensitive Detection of Ebola Virus Oligonucleotide Based on Upconversion Nanoprobe/Nanoporous Membrane System. ACS Nano 2016, 10, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bu, W.; Pan, L.; Shi, J. NIR-Triggered Anticancer Drug Delivery by Upconverting Nanoparticles with Integrated Azobenzene-Modified Mesoporous Silica. Angew. Chem. Int. Ed. 2013, 52, 4375–4379. [Google Scholar] [CrossRef] [PubMed]
- Ai, F.; Sun, T.; Xu, Z.; Wang, Z.; Kong, W.; To, M.W.; Wang, F.; Zhu, G. An Upconversion Nanoplatform for Simultaneous Photodynamic Therapy and Pt Chemotherapy to Combat Cisplatin Resistance. Dalton Trans. 2016, 45, 13052–13060. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, P.; Li, C.; Wang, Y.; Liu, Z. Upconversion Fluorescence Resonance Energy Transfer Based Biosensor for Ultrasensitive Detection of Matrix Metalloproteinase-2 in blood. Anal. Chem. 2012, 84, 1466–1473. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhu, H.; Zhang, X.; Zhu, J.; Burda, C. Rhodamine B Derivative-Functionalized Upconversion Nanoparticels for FRET-based Fe3+-sensing. Chem. Commun. 2013, 49, 7797–7799. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yoo, B.; Lee, H.; Cha, G.; Lee, H.-S.; Cho, Y.; Kim, S.Y.; Seo, H.; Lee, W.; Son, D.; et al. Ultra-Wideband Multi-Dye-Sensitized Upconverting Nanoparticles for Information Security Application. Adv. Mater. 2017, 29, 1603169–1603176. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.; Liu, L.; Ju, Q.; Liu, Y.; Zhu, H.; Li, R.; Chen, X. Time-resolved FRET Biosensor Based on Amine-Functionalized Lanthanide-Doped NaYF4 Nanocrystals. Angew. Chem. Int. Ed. 2011, 50, 6306–6310. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhou, S.; Chen, Z.; Hu, P.; Liu, Y.; Tu, D.; Zhu, H.; Li, R.; Huang, M.; Chen, X. Sub-10 nm Lanthanide-Doped CaF2 Nanoprobes for Time-Resolved Luminescent Biodetection. Angew. Chem. Int. Ed. 2013, 52, 6671–6676. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Ju, Q.; Chen, X.; Ma, R.; Chen, B.; Bai, G.; Hao, J.; Qiao, X.; Fan, X.; Wang, F. Lanthanide-Doped Energy Cascade Nanoparticles: Full Spectrum Emission by Single Wavelength Excitation. Chem. Mater. 2015, 27, 3115–3120. [Google Scholar] [CrossRef]
- Lee, S.O.; Lou, W.; Qureshi, K.M.; Mehraein-Ghomi, F.; Trump, D.L.; Gao, A.C. RNA interference targeting Stat3 inhibits growth and induces apoptosis of human prostate cancer cells. Prostate 2004, 60, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E. Stat3 as an Oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef]
- Lassmann, S.; Schuster, I.; Walch, A.; Gobel, H.; Jutting, U.; Makowiec, F.; Hopt, U.; Werner, M. STAT3 mRNA and protein expression in colorectal cancer: Effects on STAT3-inducible targets linked to cell survival and proliferation. J. Clin. Pathol. 2007, 60, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Deng, R.; Liu, X. Preparation of Core-Shell NaGdF4 Nanoparticles Doped with Luminescent Lanthanide Ions to Be Used as Upconversion-Based Probes. Nat. Protoc. 2014, 9, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.-X.; Zhang, Y.-W.; Si, R.; Yan, Z.-G.; Sun, L.-D.; You, L.-P.; Yan, C.-H. High-Quality Sodium Rare-Earth Fluoride Nanocrystals: Controlled Synthesis and Optical Properties. J. Am. Chem. Soc. 2006, 128, 6426–6436. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Sun, T.; Chen, B.; Chen, X.; Ai, F.; Zhu, X.; Li, M.; Zhang, W.; Zhu, G.; Wang, F. A General Strategy for Ligand Exchange on Upconversion Nanoparticles. Inorg. Chem. 2017, 56, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Ju, Q.; Chen, X.; Ai, F.; Peng, D.; Lin, X.; Kong, W.; Shi, P.; Zhu, G.; Feng, W. An Upconversion Nanoprobe Operating in the First Biological Window. J. Mater. Chem. B 2015, 3, 3548–3555. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, Y.; Zhang, S.; Wang, Y.; Liu, Z. Biosensing Platform Based on Fluorescence Resonance energy Transfer from Upconverting Nanocrystals to Graphene Oxide. Angew. Chem. Int. Ed. 2011, 50, 6851–6854. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Z.; Liu, Z. Upconversion fluorescence resonance energy transfer biosensor with aromatic polymer nanospheres as the lable-free energy acceptor. Anal. Chem. 2013, 85, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lu, F.; Wu, X.-C.; Zhu, J.-J. An Upconversion Fluorescent Resonant Energy Transfer Biosensor for Hepatitis B virus (HBV) DNA Hybridization Detection. Analyst 2015, 140, 7622–7628. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the nanocrystals are available from the authors. |


| Sample | Zeta-Potential/mV | ||
|---|---|---|---|
| Ligand-Free | PAA-Modified | DNA-Conjugated | |
| NaGdF4:Ce (Small) | 47.7 | −10.6 | 3.71 |
| NaGdF4:Tb (Small) | 52.5 | −23.3 | 14.2 |
| NaGdF4:Ce (Large) | 46.8 | −5.81 | 3.04 |
| NaGdF4:Tb (Large) | 48.2 | −13.7 | 15.0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, W.; Lam, C.F.; Wang, F. An All-Nanocrystal Biosensing System for In Vitro Detection of STAT3 Oligonucleotides. Molecules 2017, 22, 1085. https://doi.org/10.3390/molecules22071085
Kong W, Lam CF, Wang F. An All-Nanocrystal Biosensing System for In Vitro Detection of STAT3 Oligonucleotides. Molecules. 2017; 22(7):1085. https://doi.org/10.3390/molecules22071085
Chicago/Turabian StyleKong, Wei, Chau Fan Lam, and Feng Wang. 2017. "An All-Nanocrystal Biosensing System for In Vitro Detection of STAT3 Oligonucleotides" Molecules 22, no. 7: 1085. https://doi.org/10.3390/molecules22071085




