Neighbor Affinity-Based Core-Attachment Method to Detect Protein Complexes in Dynamic PPI Networks
Abstract
:1. Introduction
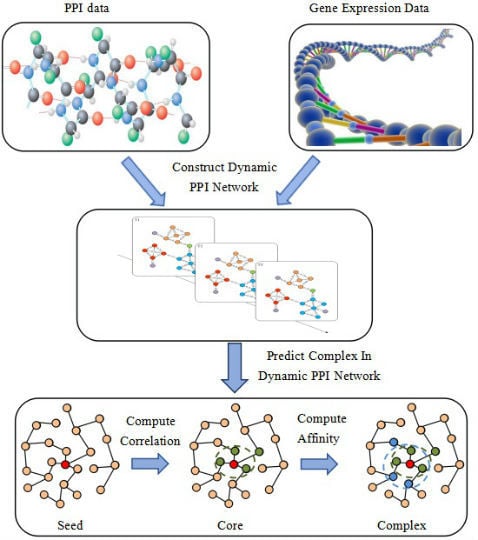
2. Method
2.1. Dynamic PPI Networks Construction
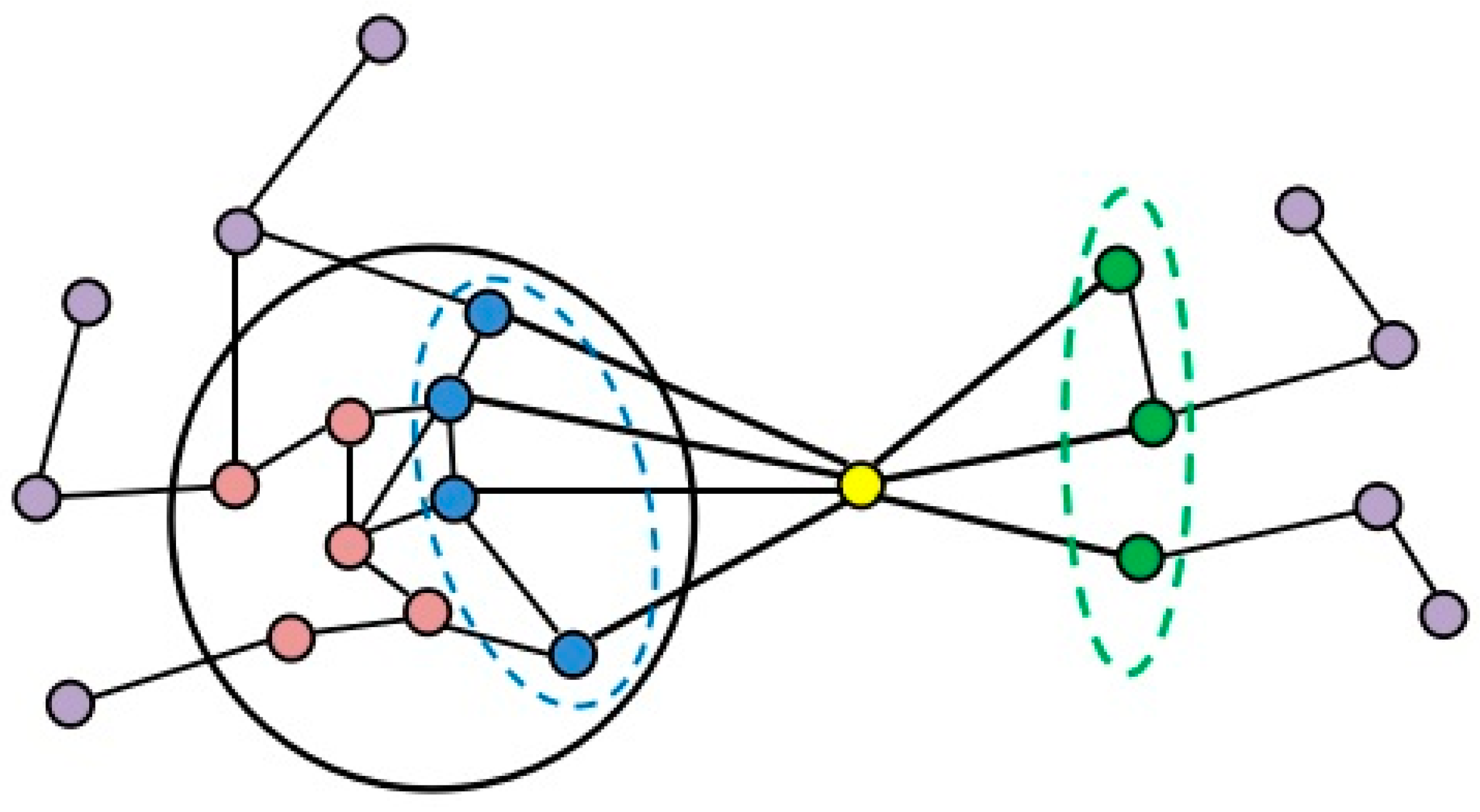
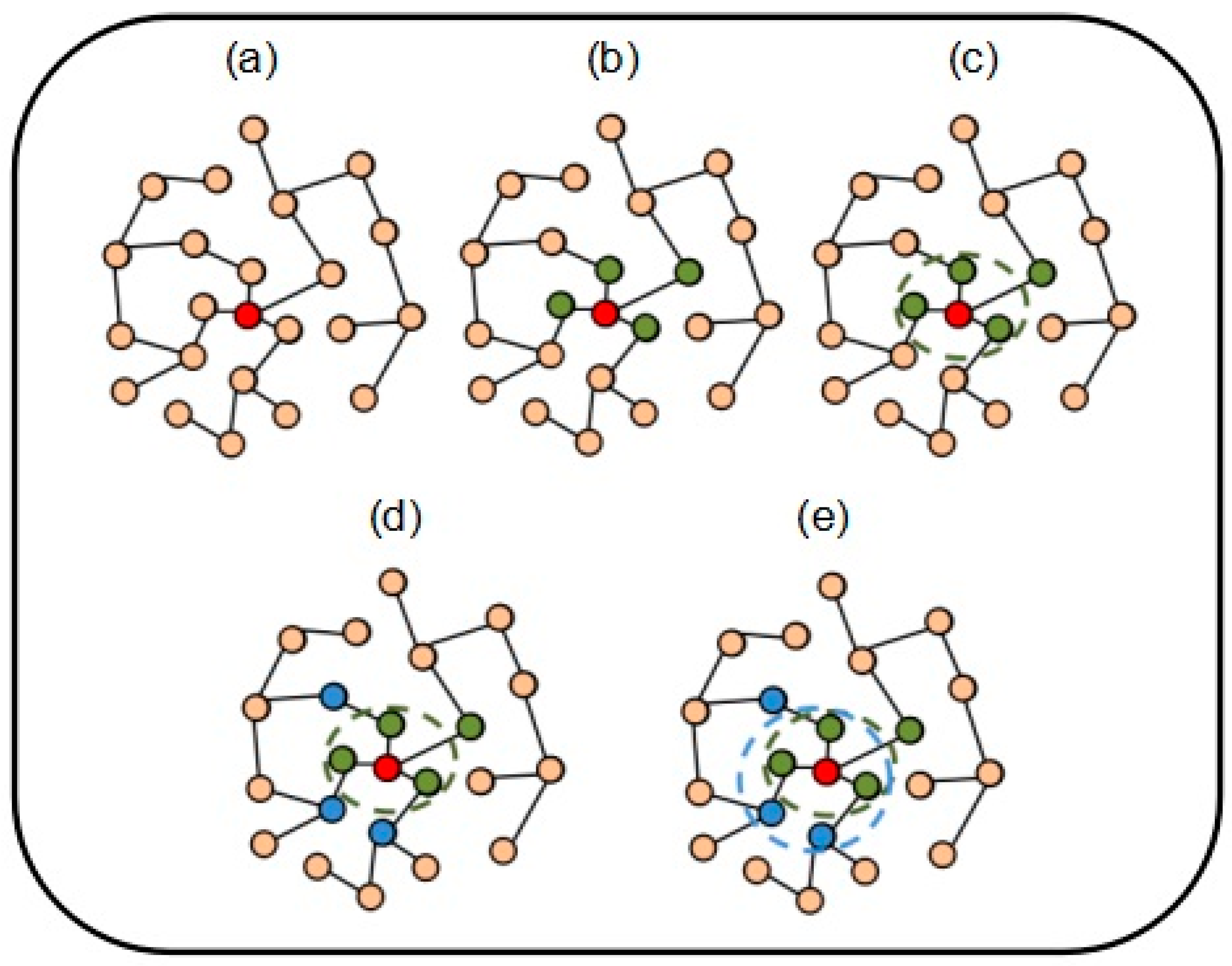
2.2. Formation Process of Attachment
2.3. NABCAM Algorithm
3. Experiments and Results
3.1. Experimental Datasets
3.2. Evaluation Criteria
3.3. Comparison with Known Complexes
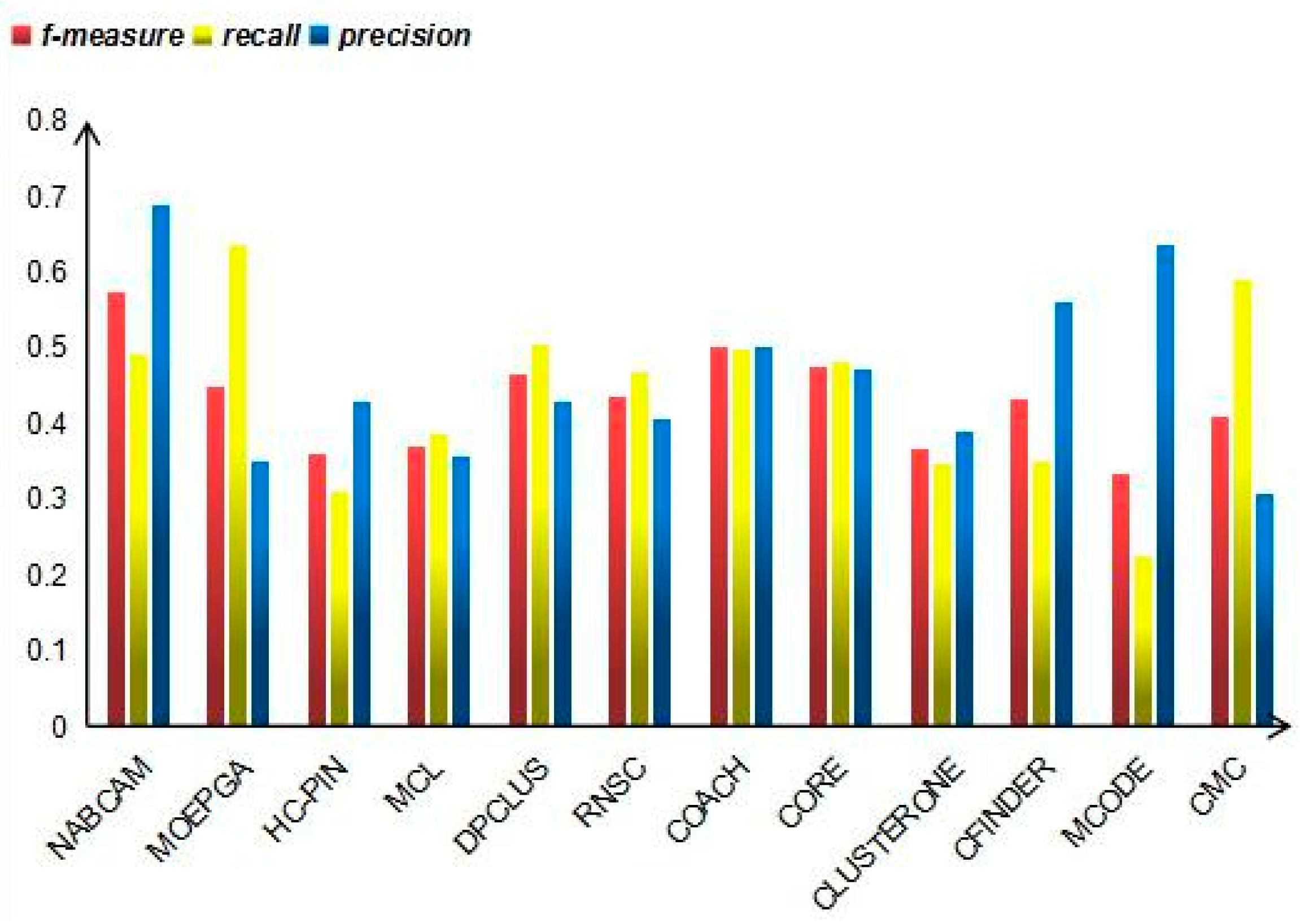
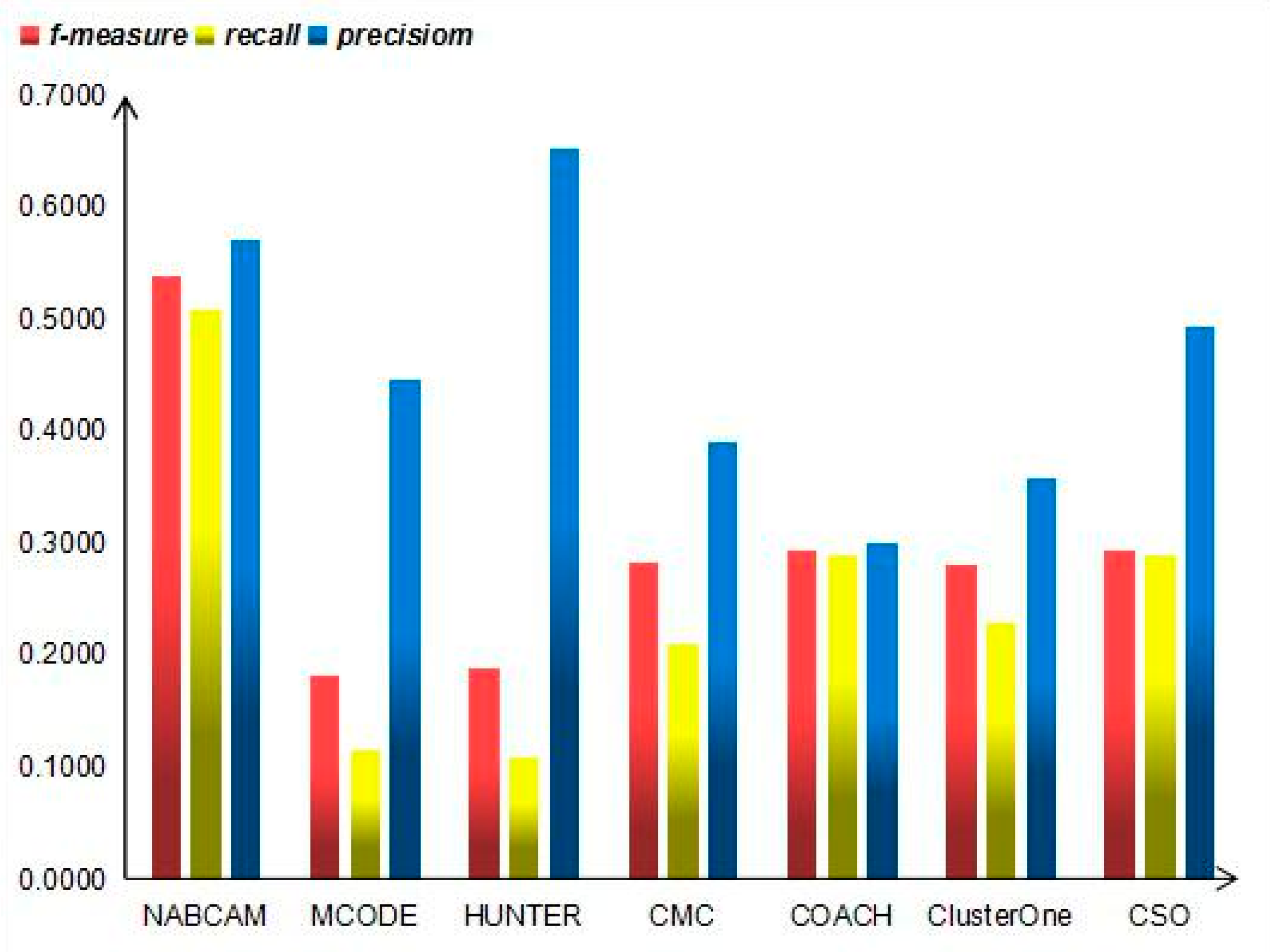
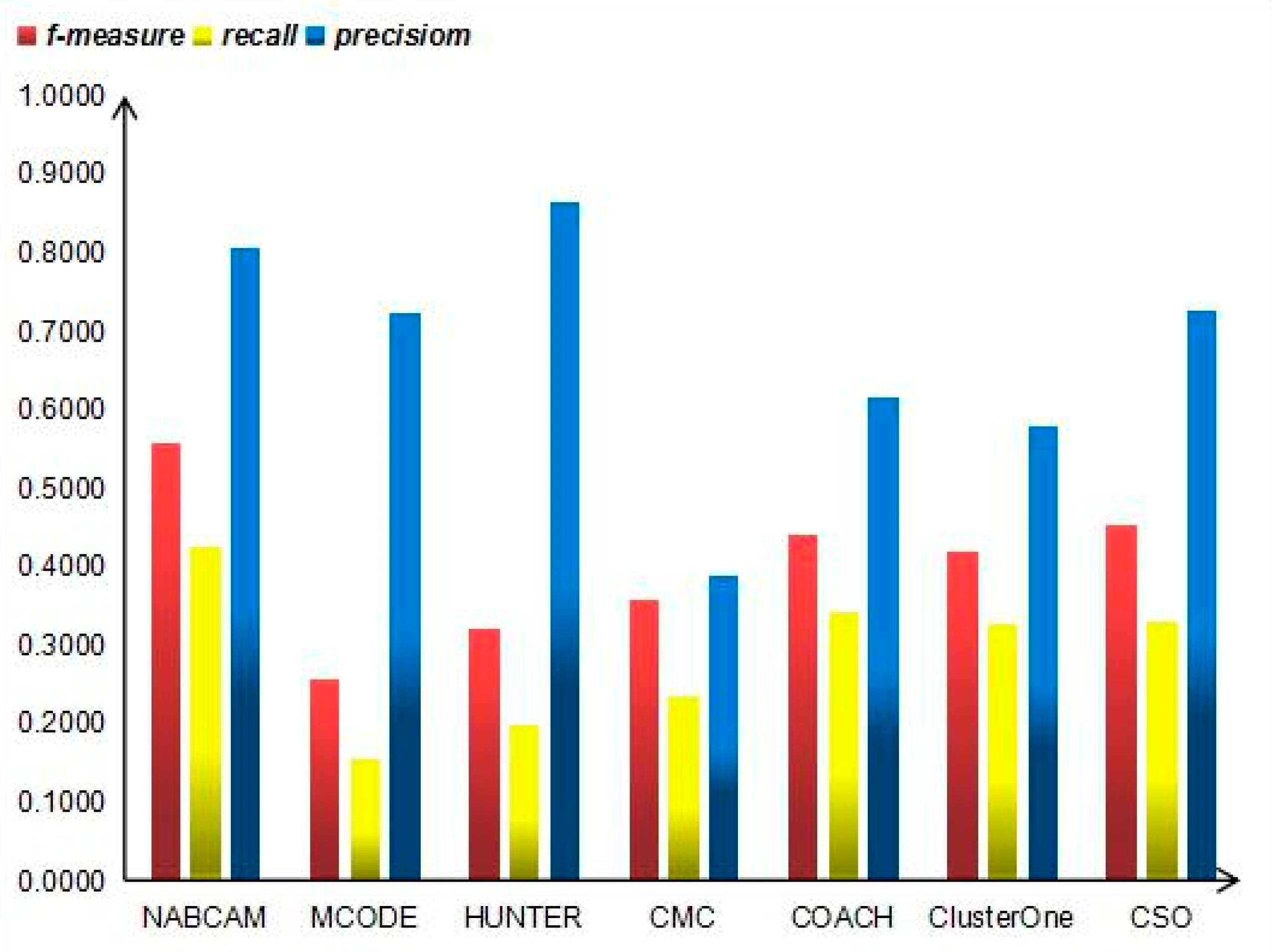
3.4. Comparison Based on Precision, Recall and F-Measure
3.5. Comparison Based on Gene Ontology (GO) Semantic
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, Y.; You, Z.; Li, X.; Chen, X.; Jiang, T.; Zhang, J. PCVMZM: Using the probabilistic classification vector machines model combined with a zernike moments descriptor to predict protein-protein interactions from protein sequences. Int. J. Mol. Sci. 2017, 18, 1029. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhang, X.; Zou, Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Brief. Bioinform. 2016, 17, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Almedia, R.M.; Acqua, S.D.; Krippahl, L.; Moura, J.J.G.; Pauleta, S.R. Predicting protein-protein interactions using bigger: Case studies. Molecules 2016, 21, 1037. [Google Scholar] [CrossRef] [PubMed]
- .Georgiou, D.N.; Karakasidis, T.E.; Megaritis, A.C. A short survey on genetic sequences, chou’s pseudo amino acid composition and its combination with fuzzy set theory. Open Bioinform. J. 2013, 7, 41–48. [Google Scholar] [CrossRef]
- Ohno, M.; Karagiannis, P.; Taniguchi, Y. Protein expression analyses at the single cell level. Molecules 2014, 19, 13932–13947. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tang, J.; Guo, F. Identification of protein-protein interactions via a novel matrix-based sequence representation model with amino acid contact information. Int. J. Mol. Sci. 2016, 17, 1623. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Wong, L.; Chua, H.N. Complex discovery from weighted PPI networks. Bioinform. 2009, 25, 1891–1897. [Google Scholar] [CrossRef] [PubMed]
- Srihari, S.; Ning, K.; Leong, H.W. MCL-CAw: A refinement of MCL for detecting yeast complexes from weighted PPI networks by incorporating core-attachment structure. BMC Bioinform. 2010, 11, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nepusz, T.; Yu, H.; Paccanaro, A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods 2012, 9, 471–472. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Fan, W.; Liu, J.; Wu, F.X. Identifying protein complexes and functional modules–from static PPI networks to dynamic PPI networks. Brief. Bioinform. 2014, 15, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Peng, X.; Li, M.; Pan, Y. Construction and application of dynamic protein interaction network based on time course gene expression data. Proteomics 2013, 13, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Bader, J. How networks change with time. Bioinformatics 2012, 28, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Ou-Yang, L.; Dai, D.Q.; Li, X.L.; Wu, M.; Zhang, X.F.; Yang, P. Detecting temporal protein complexes from dynamic protein-protein interaction networks. BMC Bioinform. 2014, 15, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, W.; Wang, J.; Wu, F.X.; Pan, Y. Identifying dynamic protein complexes based on gene expression profiles and PPI networks. BioMed Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.C.; Aloy, P.; Grandi, P.; Krause, R.; Boesche, M.; Marzioch, M.; Rau, C.; Jensen, L.J.; Bastuck, S.; Dümpelfeld, B. Proteome survey reveals modularity of the yeast cell machinery. Nature 2006, 440, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, X.; Kwoh, C.K.; Ng, S.K. A core-attachment based method to detect protein complexes in PPI networks. BMC Bioinform. 2009, 10, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Pizzuti, C.; Rombo, S. Experimental evaluation of topological-based fitness functions to detect complexes in PPI networks. In Proceedings of the 14th annual conference on Genetic and evolutionary computation, Philadelphia, PA, USA, 7–11 July 2012; pp. 193–200. [Google Scholar]
- Pan, J.; Hu, S.; Wang, H.; Zou, Q. Pagefinder: Quantitative identification of spatiotemporal pattern genes. Bioinformatics 2012, 28, 1544–1545. [Google Scholar] [CrossRef] [PubMed]
- Dezso, Z.; Oltvai, Z.N.; Barabasi, A.L. Bioinformatics analysis of experimentally determined protein complexes in the yeast Saccharomyces cerevisiae. Genome Res. 2003, 13, 2450–2454. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, J.; Huan, J.; Wu, F.X. Double-layer clustering method to predict protein complexes based on power-law distribution and protein sublocalization. J. Theor. Biol. 2016, 395, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Tang, C.; Zhang, A. Cluster analysis for gene expression data: A survey. IEEE Trans. Knowl. Data Eng. 2004, 16, 1370–1386. [Google Scholar] [CrossRef]
- King, A.D.; Pržulj, N.; Jurisica, I. Protein complex prediction via cost-based clustering. Bioinformatics 2004, 20, 3013–3020. [Google Scholar] [CrossRef] [PubMed]
- Güldener, U.; Münsterkötter, M.; Oesterheld, M.; Pagel, P.; Ruepp, A.; Mewes, H. Mpact: The MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006, 34, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Krogan, N.; Cagney, G.; Yu, H.; Zhong, G.; Guo, X.; Ignatchenko, A. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 2006, 440, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Keretsu, S.; Sarmah, R. Weighted edge based clustering to identify protein complexes in protein–protein interaction networks incorporating gene expression profile. Comput. Biol. Chem. 2016, 65, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Yi, L.; Jiang, X.; Zhao, Y.; Hu, X.; He, T.; Yang, J. Neighbor affinity based algorithm for discovering temporal protein complex from dynamic PPI network. Methods 2016, 110, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Luo, J.; Liang, C.; Wang, S.; Song, D. Moepga: A novel method to detect protein complexes in yeast protein–protein interaction networks based on multiobjective evolutionary programming genetic algorithm. Comput. Biol. Chem. 2015, 58, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, M.; Chen, J.; Pan, Y. A fast hierarchical clustering algorithm for functional modules discovery in protein interaction networks. Comput. Biol. Bioinform. 2011, 8, 607–620. [Google Scholar]
- Leal, J.P.; Enright, A.; Ouzounis, C.A. Detection of functional modules from protein interaction networks. Proteins Struct. Funct. Bioinform. 2003, 54, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Altaf-UI-Amin, M.; Shinbo, Y.; Mihara, K.; Kurokawa, K.; Kanaya, S. Development and implementation of an algorithm for detection of protein complexes in large interaction networks. BMC Bioinform. 2006, 7, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.C.; Xiang, Q.; Yiu, S.M.; Chin, F.Y. Predicting protein complexes from PPI data: A core-attachment approach. J. Comput. Biol. 2009, 16, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Adamcsek, B.; Palla, G.; Farkas, I.J.; Vicsek, T. CFinder: Locating cliques and overlapping modules in biological networks. Bioinformatics 2006, 22, 1021–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, H.; Yang, Z.; Wang, J.; Li, Y.; Xu, B. Protein complex prediction in large ontology attributed protein-protein interaction networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.; Chen, S.; Ho, C.; Ko, M.; Lin, C. A hub-attachment based method to detect functional modules from confidence-scored protein interactions and expression profiles. BMC Bioinform. 2010, 11, S25. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |



| Data | Timestamp | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DIP | Proteins | 797 | 941 | 796 | 623 | 601 | 530 | 493 | 944 | 1090 | 592 | 661 | 461 |
| Interactions | 981 | 1444 | 1188 | 745 | 750 | 646 | 573 | 1705 | 2185 | 856 | 974 | 526 | |
| MIPS | Proteins | 737 | 897 | 781 | 583 | 570 | 531 | 470 | 839 | 1014 | 523 | 616 | 402 |
| Interactions | 1097 | 1443 | 1183 | 754 | 684 | 642 | 504 | 1238 | 1637 | 878 | 1207 | 700 | |
| Krogan | Proteins | 336 | 379 | 320 | 256 | 206 | 189 | 202 | 580 | 626 | 304 | 330 | 250 |
| Interactions | 334 | 464 | 331 | 234 | 210 | 184 | 213 | 1025 | 1081 | 314 | 373 | 258 |
| Data | Algorithm | PC | <10−15 | [10−15, 10−10) | [10−10, 10−5) | [10−5, 0.01) | ≥0.01 |
|---|---|---|---|---|---|---|---|
| DIP | NABCAM | 1702 | 136 (7.99%) | 230 (13.51%) | 820 (48.18%) | 343 (20.15%) | 173 (10.16%) |
| MCL | 1053 | 19 (1.80%) | 47 (4.46%) | 183 (17.38%) | 362 (34.38%) | 442 (41.98%) | |
| CORE | 344 | 1 (0.29%) | 3 (0.87%) | 78 (22.67%) | 114 (33.14%) | 148 (43.02%) | |
| ClusterOne | 574 | 21 (3.66%) | 52 (9.06%) | 177 (30.84%) | 184 (32.06%) | 140 (24.39%) | |
| MIPS | NABCAM | 966 | 30 (3.10%) | 70 (7.25%) | 332 (34.37%) | 333 (34.47%) | 201 (20.81%) |
| MCL | 606 | 5 (0.83%) | 13 (2.15%) | 94 (15.51%) | 220 (36.30%) | 274 (45.21%) | |
| CORE | 340 | 0 (0.00%) | 4 (1.18%) | 65 (19.12%) | 107 (31.47%) | 164 (48.24%) | |
| ClusterOne | 372 | 7 (1.88%) | 16 (4.30%) | 117 (31.45%) | 126 (33.87%) | 106 (28.49%) | |
| Krogan | NABCAM | 587 | 75 (12.78%) | 75 (12.78%) | 304 (51.79%) | 108 (18.39%) | 25 (4.26%) |
| MCL | 403 | 16 (3.97%) | 43 (10.67%) | 103 (25.56%) | 119 (29.53%) | 122 (30.27%) | |
| CORE | 255 | 3 (1.18%) | 10 (3.92%) | 60 (23.53%) | 102 (40.00%) | 80 (31.37%) | |
| ClusterOne | 399 | 13 (3.26%) | 43 (10.78%) | 98 (24.56%) | 120(30.08%) | 125 (31.33%) |
| Data | ID | p-Value | Cluster Frequency | Gene Ontology Term |
|---|---|---|---|---|
| DIP | 1 | 2.45 × 10−47 | 30 out of 34 genes, 88.2% | ribosomal small subunit biogenesis |
| 2 | 4.48 × 10−38 | 22 out of 23 genes, 95.7% | mRNA splicing, via spliceosome | |
| 3 | 1.41 × 10−37 | 21 out of 21 genes, 100.0% | mRNA splicing, via spliceosome | |
| 4 | 3.88 × 10−36 | 22 out of 23 genes, 95.7% | ribosomal small subunit biogenesis | |
| 5 | 1.45 × 10−33 | 12 out of 12 genes, 100.0% | polyadenylation-dependent snoRNA 3′-end processing | |
| MIPS | 1 | 9.62 × 10−27 | 16 out of 18 genes, 88.9% | ribosomal large subunit biogenesis |
| 2 | 1.15 × 10−25 | 18 out of 25 genes, 72.0% | mitotic sister chromatid segregation | |
| 3 | 1.15 × 10−23 | 16 out of 17 genes, 94.1% | mitotic nuclear division | |
| 4 | 4.02 × 10−23 | 14 out of 16 genes, 87.5% | ribosomal large subunit biogenesis | |
| 5 | 2.69 × 10−22 | 16 out of 23 genes, 69.6% | mitotic sister chromatid segregation | |
| Krogan | 1 | 1.36 × 10−34 | 17 out of 18 genes, 94.4% | ncRNA transcription |
| 2 | 3.69 × 10−34 | 13 out of 15 genes, 86.7% | tRNA catabolic process | |
| 3 | 2.49 × 10−33 | 13 out of 14 genes, 92.9% | chromatin disassembly | |
| 4 | 3.17 × 10−32 | 13 out of 16 genes, 81.2% | exonucleolytic trimming to generate mature 3′-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | |
| 5 | 5.79 × 10−32 | 18 out of 18 genes, 100.0% | mRNA splicing, via spliceosome |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, X.; Liang, J. Neighbor Affinity-Based Core-Attachment Method to Detect Protein Complexes in Dynamic PPI Networks. Molecules 2017, 22, 1223. https://doi.org/10.3390/molecules22071223
Lei X, Liang J. Neighbor Affinity-Based Core-Attachment Method to Detect Protein Complexes in Dynamic PPI Networks. Molecules. 2017; 22(7):1223. https://doi.org/10.3390/molecules22071223
Chicago/Turabian StyleLei, Xiujuan, and Jing Liang. 2017. "Neighbor Affinity-Based Core-Attachment Method to Detect Protein Complexes in Dynamic PPI Networks" Molecules 22, no. 7: 1223. https://doi.org/10.3390/molecules22071223