Apigenin Impacts the Growth of the Gut Microbiota and Alters the Gene Expression of Enterococcus
Abstract
:1. Introduction
2. Results
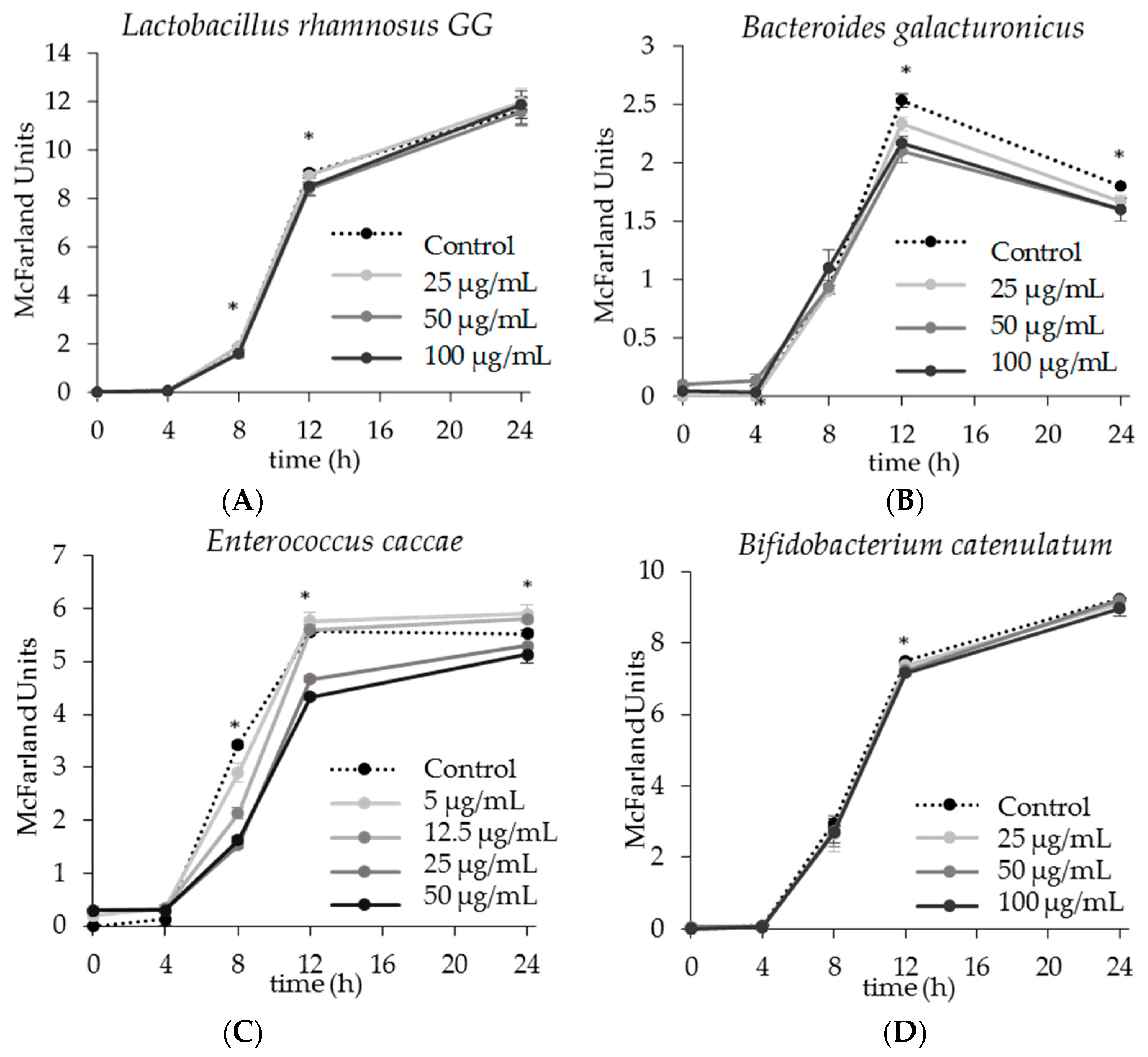
2.1. The Effect of Apigenin on Single Strain Bacterial Growth
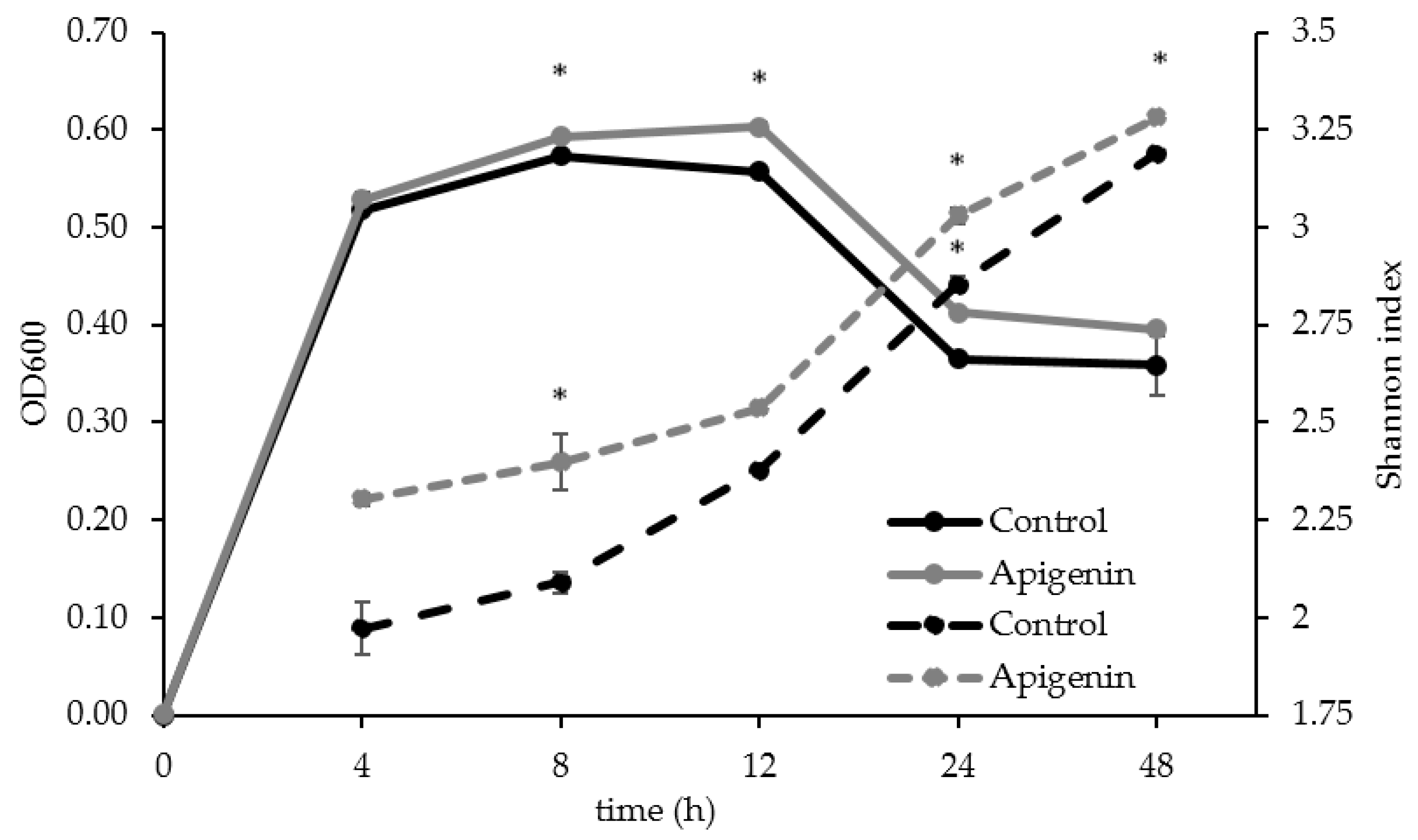
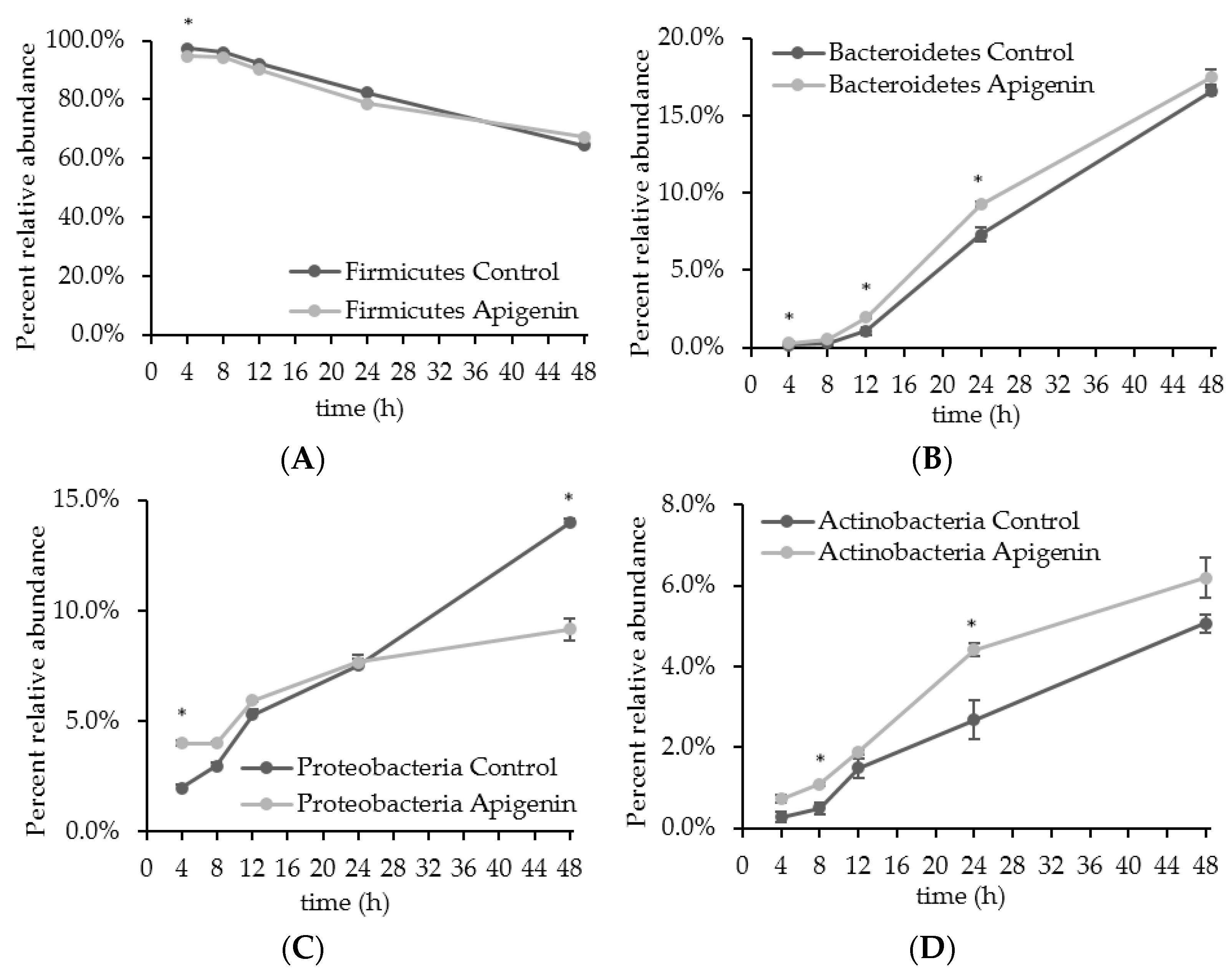
2.2. The Effect of Apigenin on a Human Gut Microbial Community In Vitro
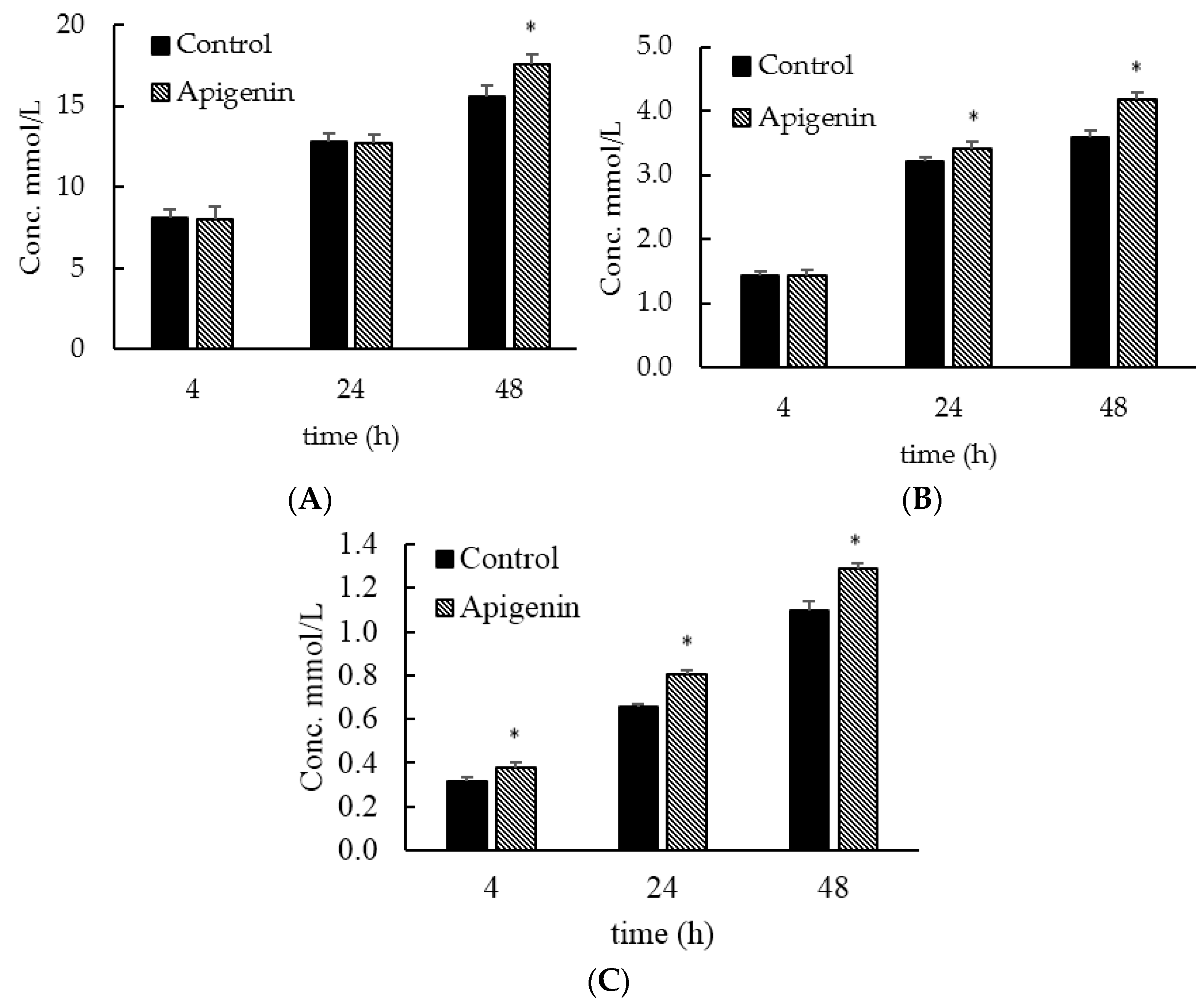
2.3. Apigenin Influences Short Chain Fatty Acids Production in a Community Setting
2.4. Apigenin-Induced Changes to the Genetic Expression Profile of Enterococcus caccae
3. Discussion
4. Materials and Methods
4.1. Media Preparation
4.2. Apigenin Stock Solutions
4.3. Single Strain Bacteria Preparation
4.4. Growth Curve Measurement of Single Strain Bacteria
4.5. Figures and Statistics for Single Strain Bacteria Growth Curve
4.6. Inoculum Preparation for Batch Culture Experiment
4.7. Batch Culture Fermentation
4.8. DNA Extraction
4.9. DNA Sequencing
4.10. Bioinformatics Processing
4.11. RNA Extraction
4.12. RNA Sequencing: rRNA Removal, cDNA Synthesis, Poly A Tailing and Blocking
4.13. RNA Sequencing Analysis
4.14. Short Chain Fatty Acid Quantification by GC-MS
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bhagwat, S.; Haytowitz, D.B.; Holden, J.M. USDA Database for the Flavonoid Content of Selected Foods Release 3.1. 2014. Available online: https://www.ars.usda.gov/ARSUserFiles/80400525/Data/Flav/Flav_R03-1.pdf (accessed on 10 July 2017).
- Lee, W.J.; Chen, W.K.; Wang, C.J.; Lin, W.L.; Tseng, T.H. Apigenin inhibits HGF-promoted invasive growth and metastasis involving blocking PI3K/Akt pathway and β4 integrin function in MDA-MB-231 breast cancer cells. Toxicol. Appl. Pharmacol. 2008, 226, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Gee, J.M.; DuPont, M.S.; Rhodes, M.J.; Johnson, I.T. Quercetin glucosides interact with the intestinal glucose transport pathway. Free Radic. Biol. Med. 1998, 25, 19–25. [Google Scholar] [CrossRef]
- Janssen, K. Dietary Non-Nutrients and Haemostasis in Humans: Effects of Salicylates, Flavonoids and Ginger; Ponsen & Looijen: Wageningen, The Netherlands, 1997. [Google Scholar]
- Mullie, P.; Clarys, P.; Deriemaeker, P.; Hebbelinck, M. Estimation of daily human intake of food flavonoids. Plant Foods Hum. Nutr. 2007, 62, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Suzaki, S.; Nishikawa, T.; Kihara, M.; Shibata, H.; Higuti, T. Phytochemical flavones isolated from Scutellaria barbata and antibacterial activity against methicillin-resistant Staphylococcus aureus. J. Ethnopharmacol. 2000, 72, 483–488. [Google Scholar] [CrossRef]
- Basile, A.; Sorbo, S.; Giordano, S.; Ricciardi, L.; Ferrara, S.; Montesano, D.; Castaldo Cobianchi, R.; Vuotto, M.L.; Ferrara, L. Antibacterial and allelopathic activity of extract from Castanea sativa leaves. Fitoterapia 2000, 71, 110–116. [Google Scholar] [CrossRef]
- Nayaka, H.B.; Londonkar, R.L.; Umesh, M.K.; Tukappa, A. Antibacterial attributes of apigenin, isolated from Portulaca oleracea L. Int. J. Bacteriol. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rigano, D.; Formisano, C.; Basile, A.; Lavitola, A.; Senatore, F.; Rosselli, S.; Bruno, M. Antibacterial activity of flavonoids and phenylpropanoids from Marrubium globosum ssp. libanoticum. Phytother. Res. 2007, 21, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Martini, N.D.; Katerere, D.R.P.; Eloff, J.N. Biological activity of five antibacterial flavonoids from Combretum erythrophyllum (Combretaceae). J. Ethnopharmacol. 2004, 93, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yi, Z.B.; Liang, Y.Z. Validate antibacterial mode and find main bioactive components of traditional Chinese medicine Aquilegia oxysepala. Bioorg. Med. Chem. Lett. 2007, 17, 1855–1859. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Shukla, S.; Gupta, S. Apigenin and cancer chemoprevention: Progress, potential and promise (Review). Int. J. Oncol. 2007, 30, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, C.; Batra, S.; Vargo, M.A.; Voss, O.H.; Gavrilin, M.A.; Wewers, M.D.; Guttridge, D.C.; Grotewold, E.; Doseff, A.I. Apigenin blocks lipopolysaccharide-induced lethality in vivo and proinflammatory cytokines expression by inactivating NF-κB through the suppression of p65 phosphorylation. J. Immunol. 2007, 179, 7121–7127. [Google Scholar] [CrossRef] [PubMed]
- Romanová, D.; Vachálková, A.; Cipák, L.; Ovesná, Z.; Rauko, P. Study of antioxidant effect of apigenin, luteolin and quercetin by DNA protective method. Neoplasma 2001, 48, 104–107. [Google Scholar] [PubMed]
- Shukla, S.; Gupta, S. Apigenin: A promising molecule for cancer prevention. Pharm. Res. 2010, 27, 962–978. [Google Scholar] [CrossRef] [PubMed]
- Cardona, F.; Andrés-Lacueva, C.; Tulipani, S.; Tinahones, F.J.; Queipo-Ortuño, M.I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 2013, 24, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, L.P.; Lu, X.Y.; Jiang, H.D.; Zeng, S. Absorption and excretion of luteolin and apigenin in rats after oral administration of Chrysanthemum morifolium extract. J. Agric. Food Chem. 2007, 55, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Gradolatto, A.; Basly, J.-P.; Berges, R.; Teyssier, C.; Chagnon, M.-C.; Siess, M.-H.; Canivenc-Lavier, M.-C. Pharmacokinetics and metabolism of apigenin in female and male rats after a single oral administration. Drug Metab. Dispos. 2005, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Schoefer, L.; Mohan, R.; Schwiertz, A.; Braune, A.; Blaut, M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Microbiology 2003, 69, 5849–5854. [Google Scholar] [CrossRef]
- Hanske, L.; Loh, G.; Sczesny, S.; Blaut, M.; Braune, A. The bioavailability of apigenin-7-glucoside is influenced by human intestinal microbiota in rats. J. Nutr. 2009, 139, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Ulanowska, K.; Tkaczyk, A.; Konopa, G.; Wȩgrzyn, G. Differential antibacterial activity of genistein arising from global inhibition of DNA, RNA and protein synthesis in some bacterial strains. Arch. Microbiol. 2006, 184, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Hamilton, V.E.S.; Lamb, A.J. Assessment of the antibacterial activity of selected flavonoids and consideration of discrepancies between previous reports. Microbiol. Res. 2003, 158, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Koskenniemi, K.; Laakso, K.; Koponen, J.; Kankainen, M.; Greco, D.; Auvinen, P.; Savijoki, K.; Nyman, T.A.; Surakka, A.; Salusjarvi, T.; et al. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell. Proteom. 2011, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Firrman, J.; Liu, L.; Zhang, L.; Argoty, G.A.; Wang, M.; Tomasula, P.; Kobori, M.; Pontious, S.; Xiao, W. The effect of quercetin on genetic expression of the commensal gut microbes Bifidobacterium catenulatum, Enterococcus caccae and Ruminococcus gauvreauii. Anaerobe 2016, 42, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus diversity, origins in nature, and gut colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 1–59. [Google Scholar]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.; Turnbaugh, P.; Klein, S.; Gordon, J. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Van Dorsten, F.A.; Peters, S.; Gross, G.; Gomez-Roldan, V.; Klinkenberg, M.; De Vos, R.C.; Vaughan, E.E.; Van Duynhoven, J.P.; Possemiers, S.; Van de Wiele, T.; et al. Gut microbial metabolism of polyphenols from black tea and red wine/grape juice is source-specific and colon-region dependent. J. Agric. Food Chem. 2012, 60, 11331–11342. [Google Scholar] [CrossRef] [PubMed]
- Burger-van Paassen, N.; Vincent, A.; Puiman, P.J.; van der Sluis, M.; Bouma, J.; Boehm, G.; van Goudoever, J.B.; van Seuningen, I.; Renes, I.B. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: Implications for epithelial protection. Biochem. J. 2009, 420, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, L.E.; Koetsier, M.A.; van Deventer, S.J.; van Tol, E.A. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.-R. Gut flora in health and disease. Lancet 2003, 360, 512–519. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Le Bourgeois, P.; Bugarel, M.; Campo, N.; Daveran-Mingot, M.-L.; Labonté, J.; Lanfranchi, D.; Lautier, T.; Pagès, C.; Ritzenthaler, P. The unconventional Xer recombination machinery of Streptococci/Lactococci. PLoS Genet. 2007, 3, e117. [Google Scholar] [CrossRef] [PubMed]
- Crowley, D.J.; Boubriak, I.; Berquist, B.R.; Clark, M.; Richard, E.; Sullivan, L.; DasSarma, S.; McCready, S. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2006, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Sambir, M.; Ivanova, L.B.; Bryksin, A.V.; Godfrey, H.P.; Cabello, F.C. Functional analysis of Borrelia burgdorferi uvrA in DNA damage protection. FEMS Microbiol. Lett. 2011, 317, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Grinberg, I.; Shteinberg, T.; Gorovitz, B.; Aharonowitz, Y.; Cohen, G.; Borovok, I. The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J. Bacteriol. 2006, 188, 7635–7644. [Google Scholar] [CrossRef] [PubMed]
- Truglio, J.J.; Croteau, D.L.; Van Houten, B.; Kisker, C. Prokaryotic nucleotide excision repair: The UvrABC System. Chem. Rev. 2006, 106, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-G.; Jung, M.-C.; Song, H.; Jeong, K.-W.; Bang, E.; Hwang, G.-S.; Kim, Y. Novel structural components contribute to the high thermal stability of acyl carrier protein from Enterococcus faecalis. J. Biol. Chem. 2016, 291, 1692–1702. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Zhu, L.; Wang, H.; Cronan, J.E. Inefficient translation renders the Enterococcus faecalis fabk enoyl-acyl carrier protein reductase phenotypically cryptic. J. Bacteriol. 2014, 196, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Nishida, I.; Kawaguchi, A.; Yamada, M. Effect of thiolactomycin on the individual enzymes of the fatty acid synthase system in Escherichia coli. J. Biochem. 1986, 99, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, F.; Ogawa, Y.; Takeuchi, K.; Suzuki, T.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. A homologue of the 3-oxoacyl-(acyl carrier protein) synthase III gene located in the glycosylation island of Pseudomonas syringae pv. tabaci regulates virulence factors via N-acyl homoserine lactone and fatty acid synthesis. J. Bacteriol. 2006, 188, 8376–8384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumariya, R.; Sood, S.K.; Rajput, Y.S.; Garsa, A.K. Gradual pediocin PA-1 resistance in Enterococcus faecalis confers cross-protection to diverse pore-forming cationic antimicrobial peptides displaying changes in cell wall and mannose PTS expression. Ann. Microbiol. 2015, 65, 721–732. [Google Scholar] [CrossRef]
- White, S.W.; Zheng, J.; Zhang, Y.-M.; Rock, C.O. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Panesso, D.; Mishra, N.N.; Mileykovskaya, E.; Guan, Z.; Munita, J.M.; Reyes, J.; Diaz, L.; Weinstock, G.M.; Murray, B.E.; et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Panesso, D.; McGrath, D.M.; Qin, X.; Mojica, M.F.; Miller, C.; Diaz, L.; Tran, T.T.; Rincon, S.; Barbu, E.M.; et al. Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 2011, 365, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Morath, S.; Stadelmaier, A.; Geyer, A.; Schmidt, R.R.; Hartung, T. Synthetic lipoteichoic acid from Staphylococcus aureus is a potent stimulus of cytokine release. J. Exp. Med. 2002, 195, 1635–1640. [Google Scholar] [CrossRef] [PubMed]
- Mohamadzadeh, M.; Pfeiler, E.A.; Brown, J.B.; Zadeh, M.; Gramarossa, M.; Managlia, E.; Bere, P.; Sarraj, B.; Khan, M.W.; Pakanati, K.C.; et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. USA 2011, 108, 4623–4630. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.Y.; Liu, Y.C.; Chang, C.Y.; Huang, C.J.; Chiu, Y.H.; Huang, C.M.; Hsu, N.S.; Lin, K.H.; Wu, C.J.; Tsai, M.D.; et al. Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant Enterococcus. J. Am. Chem. Soc. 2014, 136, 10989–10995. [Google Scholar] [CrossRef] [PubMed]
- Goffin, C.; Ghuysen, J.-M. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: Presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 2002, 66, 702–738. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R.A.; Harry, E.J.; Errington, J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 2000, 35, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Layec, S.; Decaris, B.; Leblond-Bourget, N. Diversity of Firmicutes peptidoglycan hydrolases and specificities of those involved in daughter cell separation. Res. Microbiol. 2008, 159, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Helmann, J.D. Reducing the level of undecaprenyl pyrophosphate synthase has complex effects on susceptibility to cell wall antibiotics. Antimicrob. Agents Chemother. 2013, 57, 4267–4275. [Google Scholar] [CrossRef] [PubMed]
- Perera, I.C.; Grove, A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2010, 2, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Zuber, P. Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 2009, 63, 575–597. [Google Scholar] [CrossRef] [PubMed]
- Zuber, P. Function and control of the Spx-family of proteins within the bacterial stress response. Briefs Microbiol. 2013, 8, 1–35. [Google Scholar]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Alcántara, C.; Zúniga, M. Proteomic and transcriptomic analysis of the response to bile stress of Lactobacillus casei BL23. Microbiology 2012, 158, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Rechner, A.R.; Smith, M.A.; Kuhnle, G.; Gibson, G.R.; Debnam, E.S.; Srai, S.K.S.; Moore, K.P.; Rice-Evans, C.A. Colonic metabolism of dietary polyphenols: Influence of structure on microbial fermentation products. Free Radic. Biol. Med. 2004, 36, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Lewis, J.D.; Hoffmann, C.; Chen, Y.-Y.; Knight, R.; Bittinger, K.; Hwang, J.; Chen, J.; Berkowsky, R.; Nessel, L.; et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- McKenna, P.; Hoffmann, C.; Minkah, N.; Aye, P.P.; Lackner, A.; Liu, Z.; Lozupone, C.A.; Hamady, M.; Knight, R.; Bushman, F.D. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008, 4, e20. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Hofstaedter, C.E.; Zhao, C.; Mattei, L.; Tanes, C.; Clarke, E.; Lauder, A.; Sherrill-Mix, S.; Chehoud, C.; Kelsen, J.; et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples are not available. |


| (A) Enterococcus caccae Genes Upregulated in Response to Apigenin | |||
|---|---|---|---|
| Gene | Description | Fold ▲ | Description of Function |
| UC7_RS11115 | hypothetical protein | 7.0 | Unknown |
| UC7_RS15590 | hypothetical protein | 5.9 | Unknown |
| UC7_RS11110 | glycerophosphoryl diester phosphodiesterase | 2.9 | Phospholipid metabolism |
| UC7_RS11955 | tyrosine recombinase XerC | 2.8 | DNA dimer resolution |
| UC7_RS11545 | hypothetical protein | 2.7 | Unknown |
| UC7_RS16330 | acyl carrier protein | 2.6 | Fatty acid biosynthesis |
| UC7_RS16340 | MarR family transcriptional regulator | 2.5 | Stress response |
| UC7_RS13900 | glycine cleavage system protein H | 2.4 | Glycine degradation |
| UC7_RS11945 | ATP-dependent protease ATPase subunit HslU | 2.3 | Protein quality control |
| UC7_RS16335 | 3-oxoacyl-ACP synthase III | 2.3 | Type II fatty acid synthesis |
| UC7_RS13125 | hypothetical protein | 2.2 | Unknown |
| UC7_RS11950 | ATP-dependent protease subunit HslV | 2.2 | Protein quality control |
| UC7_RS14535 | chaperonin | 2.2 | Protein folding |
| UC7_RS11290 | phosphoglycerol transferase | 2.1 | Lipoteichoic acid biosynthesis |
| UC7_RS15845 | hypothetical protein | 2.1 | Unknown |
| UC7_RS13905 | spx/MgsR family transcriptional regulator | 2.1 | Stress response |
| UC7_RS11940 | GTP-sensing transcriptional pleiotropic repressor CodY | 2.1 | Transcriptional regulation |
| UC7_RS13910 | cell cycle protein FtsW | 2.1 | Cell division |
| UC7_RS12345 | CBS domain-containing protein | 2.1 | metabolism |
| UC7_RS13095 | N5-carboxyaminoimidazole ribonucleotide mutase | 2.1 | Purine synthesis |
| UC7_RS15130 | acyltransferase | 2.1 | Peptidoglycan synthesis |
| UC7_RS12825 | phenazine biosynthesis protein PhzF | 2.0 | Phenazine biosynthesis |
| UC7_RS13565 | hypothetical protein | 2.0 | Unknown |
| UC7_RS15200 | excinuclease ABC subunit B | 2.0 | DNA repair |
| UC7_RS13270 | neutral zinc metallopeptidase | 1.9 | Extracellular metabolism |
| UC7_RS14315 | magnesium-transporting ATPase | 1.9 | Magnesium transportation |
| UC7_RS13560 | diaminopimelate dehydrogenase | 1.8 | Lysine biosynthesis |
| UC7_RS11285 | hypothetical protein | 1.8 | Unknown |
| UC7_RS12620 | undecaprenyl pyrophosphate synthase | 1.8 | Peptidoglycan biosynthesis |
| UC7_RS15260 | hypothetical protein | 1.7 | Unknown |
| UC7_RS16325 | enoyl-ACP reductase II | 1.7 | Fatty acid biosynthesis |
| UC7_RS12170 | hypothetical protein | 1.7 | Unknown |
| UC7_RS11935 | aldolase 1 epimerase LacX | 1.7 | Carbohydrate metabolism |
| UC7_RS11340 | cold-shock protein | 1.7 | Stress response |
| UC7_RS15195 | UvrABC system protein A | 1.7 | DNA repair |
| UC7_RS13110 | hypothetical protein | 1.7 | Unknown |
| UC7_RS13815 | hypothetical protein | 1.7 | Unknown |
| UC7_RS13990 | hypothetical protein | 1.7 | Unknown |
| UC7_RS13115 | N-acetylmuramoyl-l-alanine amidase | 1.7 | Cell separation |
| UC7_RS16345 | hypothetical protein | 1.7 | Unknown |
| UC7_RS12340 | 2_3_4_5-tetrahydropyridine-2_6-dicarboxylate N-acetyltransferase | 1.7 | Lysine biosynthesis |
| UC7_RS13365 | NrdR family transcriptional regulator | 1.7 | DNA synthesis and repair |
| UC7_RS14530 | molecular chaperone GroEL | 1.7 | Protein folding |
| UC7_RS14300 | GNAT family acetyltransferase | 1.6 | Transcriptional regulation |
| UC7_RS16190 | penicillin-binding protein 2B | 1.6 | Cell wall synthesis |
| UC7_RS13090 | phosphoribosylaminoimidazole carboxylase ATPase subunit | 1.6 | Purine and pyrimidine ribonucleotide biosynthesis |
| UC7_RS15255 | hypothetical protein | 1.6 | Unknown |
| UC7_RS13120 | hypothetical protein | 1.6 | Unknown |
| UC7_RS13335 | chaperone protein ClpB | 1.6 | Protein folding |
| UC7_RS14685 | DNA-binding response regulator | 1.6 | Transcriptional regulation |
| UC7_RS11985 | S26 family signal peptidase | 1.6 | Protein maturation |
| UC7_RS12190 | elongation factor Tu | 1.6 | Protein synthesis |
| UC7_RS15900 | d-alanine-poly(phosphoribitol) ligase subunit 2 | 1.6 | Cell wall/membrane metabolism |
| UC7_RS16320 | malonyl CoA-ACP transacylase | 1.5 | Fatty acid and polyketide synthesis |
| UC7_RS13160 | penicillin-binding protein | 1.5 | Cell wall synthesis |
| UC7_RS15715 | 50S ribosomal protein L10 | 1.5 | Protein translation |
| UC7_RS15600 | phosphocarrier protein HPr | 1.5 | Carbohydrate phosphorylation |
| UC7_RS13265 | hypothetical protein | 1.5 | Unknown |
| UC7_RS15250 | hypothetical protein | 1.5 | Unknown |
| UC7_RS12410 | signal peptidase I | 1.5 | Protein maturation |
| (B) Enterococcus caccae Genes Downregulated in Response to Apigenin | |||
| Gene | Description | Fold ▼ | Description of Function |
| UC7_RS16160 | hypothetical protein | 2.4 | Unknown |
| UC7_RS16570 | hypothetical protein | 2.2 | Unknown |
| UC7_RS14780 | hypothetical protein | 2.0 | Unknown |
| UC7_RS13490 | Lsa family ABC-F type ribosomal protection protein | 2.0 | Ribosomal protection |
| UC7_RS14785 | hypothetical protein | 1.9 | Unknown |
| UC7_RS11535 | hypothetical protein | 1.9 | Unknown |
| UC7_RS15700 | general stress protein GlsB | 1.9 | Stress response |
| UC7_RS14775 | hypothetical protein | 1.8 | Unknown |
| UC7_RS15645 | hypothetical protein | 1.8 | Unknown |
| UC7_RS16575 | hypothetical protein | 1.8 | Unknown |
| UC7_RS16225 | WxL domain surface protein | 1.7 | Surface protein binding |
| UC7_RS13920 | hypothetical protein | 1.7 | Unknown |
| UC7_RS15555 | MocD family protein | 1.7 | Catabolism of mannopine |
| UC7_RS13450 | hypothetical protein | 1.7 | Unknown |
| UC7_RS16165 | hypothetical protein | 1.7 | Unknown |
| UC7_RS13865 | SufS family cysteine desulfurase | 1.6 | Fe-S protein biosynthesis |
| UC7_RS13860 | iron-sulfur cluster assembly scaffold protein | 1.6 | Iron-sulfur cluster formation |
| UC7_RS14795 | deoxycytidine triphosphate deaminase | 1.6 | Nucleotide metabolism |
| UC7_RS13855 | FeS assembly protein SufB | 1.6 | Iron-sulfur cluster formation |
| UC7_RS13455 | hypothetical protein | 1.6 | Unknown |
| UC7_RS11040 | hypothetical protein | 1.6 | Unknown |
| UC7_RS12130 | hypothetical protein | 1.6 | Unknown |
| UC7_RS13465 | hypothetical protein | 1.5 | Unknown |
| UC7_RS12125 | hypothetical protein | 1.5 | Unknown |
| UC7_RS14270 | ATP synthase subunit C | 1.5 | ATP synthesis |
| UC7_RS16230 | hypothetical protein | 1.5 | Unknown |
| UC7_RS16220 | hypothetical protein | 1.5 | Unknown |
| UC7_RS11885 | cardiolipin synthase | 1.5 | Phospholipid biosynthesis |
| UC7_RS14805 | ATP-grasp domain-containing protein | 1.5 | metabolism |
| UC7_RS15830 | thioredoxin-disulfide reductase | 1.5 | Redox regulation |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Firrman, J.; Zhang, L.; Arango-Argoty, G.; Tomasula, P.; Liu, L.; Xiao, W.; Yam, K. Apigenin Impacts the Growth of the Gut Microbiota and Alters the Gene Expression of Enterococcus. Molecules 2017, 22, 1292. https://doi.org/10.3390/molecules22081292
Wang M, Firrman J, Zhang L, Arango-Argoty G, Tomasula P, Liu L, Xiao W, Yam K. Apigenin Impacts the Growth of the Gut Microbiota and Alters the Gene Expression of Enterococcus. Molecules. 2017; 22(8):1292. https://doi.org/10.3390/molecules22081292
Chicago/Turabian StyleWang, Minqian, Jenni Firrman, Liqing Zhang, Gustavo Arango-Argoty, Peggy Tomasula, LinShu Liu, Weidong Xiao, and Kit Yam. 2017. "Apigenin Impacts the Growth of the Gut Microbiota and Alters the Gene Expression of Enterococcus" Molecules 22, no. 8: 1292. https://doi.org/10.3390/molecules22081292





