2.1. Elucidating the Effects of L208A Substitution via Molecular Dynamic Simulation and Protein-Substrate Docking
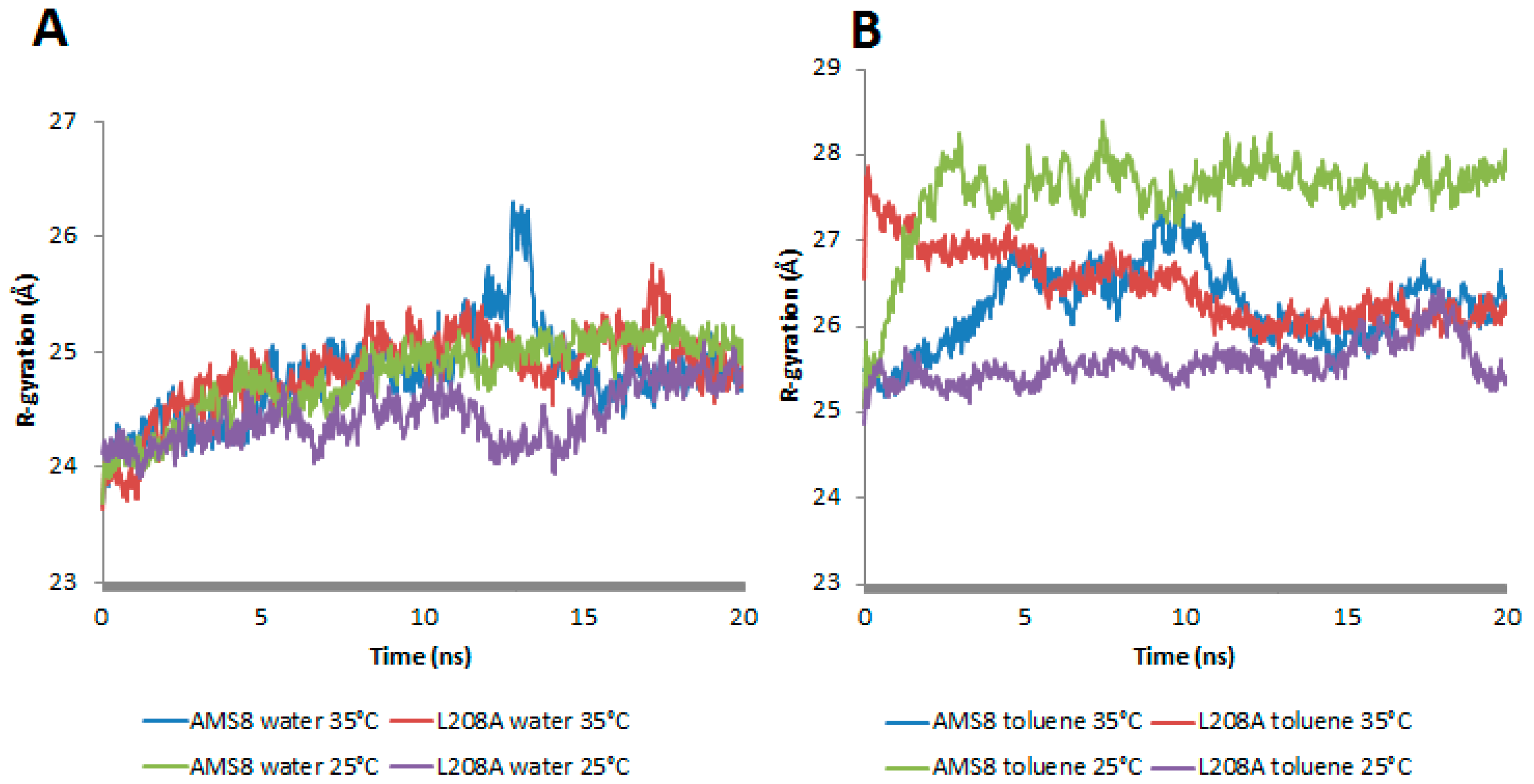
In-silico analysis of mutant L208A was studied on its molecular basis of stability in the presence of a non-polar solvent, toluene, and at different temperatures. Previously, AMS8 lipase was observed to permit “interfacial activation” in the presence of non-polar solvents. In the presence of toluene, the geometrical properties of WT AMS8 lipase showed a prominent change in terms of the increase in radius of gyration (Rg) and total accessible surface (
Figure 1B and
Figure 2B). Contrary to water simulation, the WT lipase was more rigid in movement and did not exhibit any major structural changes when the temperature was increased (
Figure 1A and
Figure 2B).
In general, mutant L208A radius of gyration was stable and sustained at 24.5 Å throughout water-simulation at both 25 °C and 35 °C. As in the presence of toluene, rapid movements of the water molecules that surround the enzyme promote irregularities in its compactness after just 1.5 ns at 25 and 35 °C. It was noteworthy that at 25 °C, L208A lipase exhibited a gradual increase in Rg starting from 25.05 Å to reach the highest value of 26.2 Å while at 35 °C, L208A radius of gyration had a sharp increase from 26.5 Å to 27.8 Å up to 1.6 ns, but finally moved back to its normal form at 25 Å after much extended simulation as shown in
Figure 1B. Hence, the mutant L208A lipase could prevent its structure from losing its native compactness in the presence of toluene at moderately high temperature (35 °C). It was believed that the improvement of compactness on L208A lipase at 35 °C has something to do with the active movement of toluene that surrounds the enzyme due to its lower volatility (higher boiling point) as compared to water molecules. Following this result, L208A lipase was more likely to favour a slight increase in temperature as shown by its structure compactness in toluene.
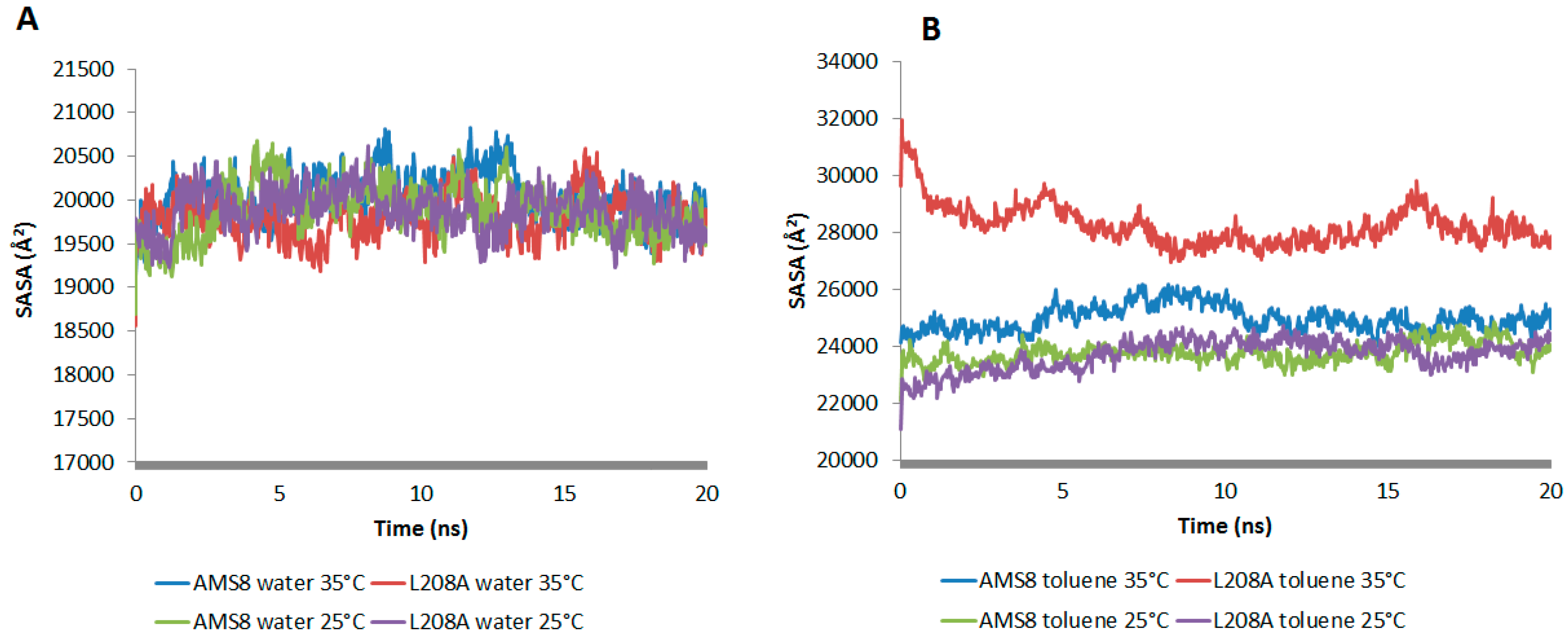
The accessible surface areas of atoms are correlated with their hydrophobicity and folding process. The folding process is usually accompanied by a significant decrease in SASA value. In water simulation, both WT AMS8 and L208A lipase showed to have similar SASA pattern at 25 °C and 35 °C, which is indicative of having no difference in the folding process (
Figure 2A). However since toluene was added into the simulation, the average value of SASA increased by 1 to 1.5 times (
Figure 2B). Even though both WT and L208A lipases exhibited an increase in SASA when simulated at 35 °C, the latter one was shown to have undergone maximum local rearrangements that modify the energy interactions with the solvent which could influence the state of protein folding itself.
There are higher chances of surface area contact between lipases in toluene as compared to water. Although the area function is not specific to the structures of proteins, the derivation was motivated by the need for a computationally feasible simulation of the hydrophobic effect in proteins [
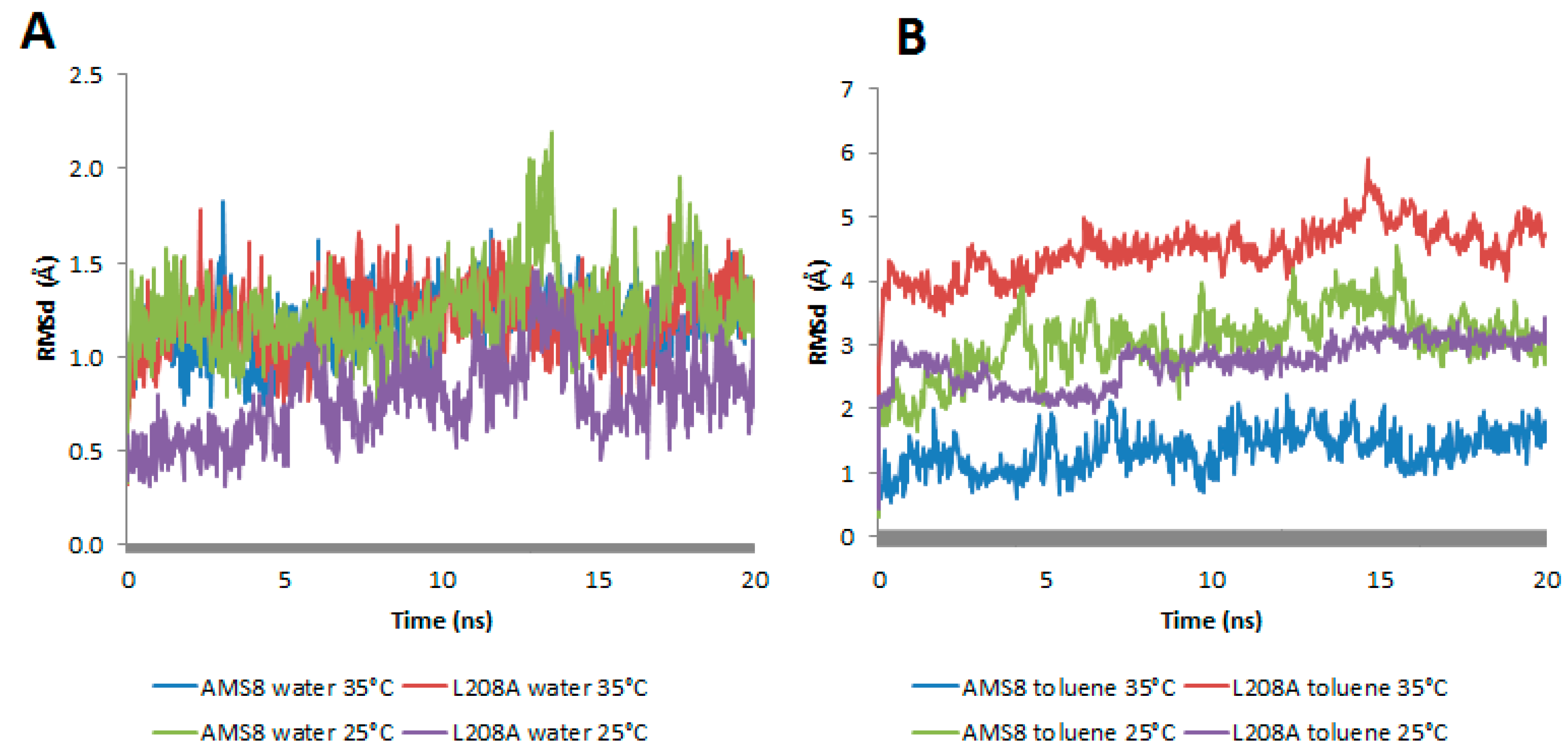
13]. Therefore, lid 2, a region known for its hydrophobicity interactions, was targeted and compared for its root-mean-square deviations (RMSd) at both conditions. In general, both WT and mutant L208A lipases displayed an average distance of 1–1.5 Å in water simulation at 25 °C and 35 °C (
Figure 3A). This analysis revealed that water stabilizes the lid at both temperatures. A change in lid stability of mutant L208A lipase could be found prior to temperature changes in toluene as shown in
Figure 3B. The difference in thermal condition happens to change mutant L208A lid stability at 35 °C following the escalating RMSd value. As a result of this, mutant L208A was predicted to have experienced a disturbance in lid stability and the possibility of undergoing thermal denaturation, while at lower temperature (25 °C), lid 2 of L208A lipase was shown to stabilize and reach equilibrium with the RMSd value of the wild-type strain. WT AMS8 lipase exhibited a further decline in RMSd values when simulated at 35 °C as shown in
Figure 3B and this effect could be associated with the thermolability as high temperature that abolishes the stability of cold-active lipase. Based on molecular dynamics, a large difference of RMSd between WT and L208A lipases had raised concerns about the potential impact of a single mutation at the nucleophilic elbow region on its ability to alter the lid stability at different temperature settings.
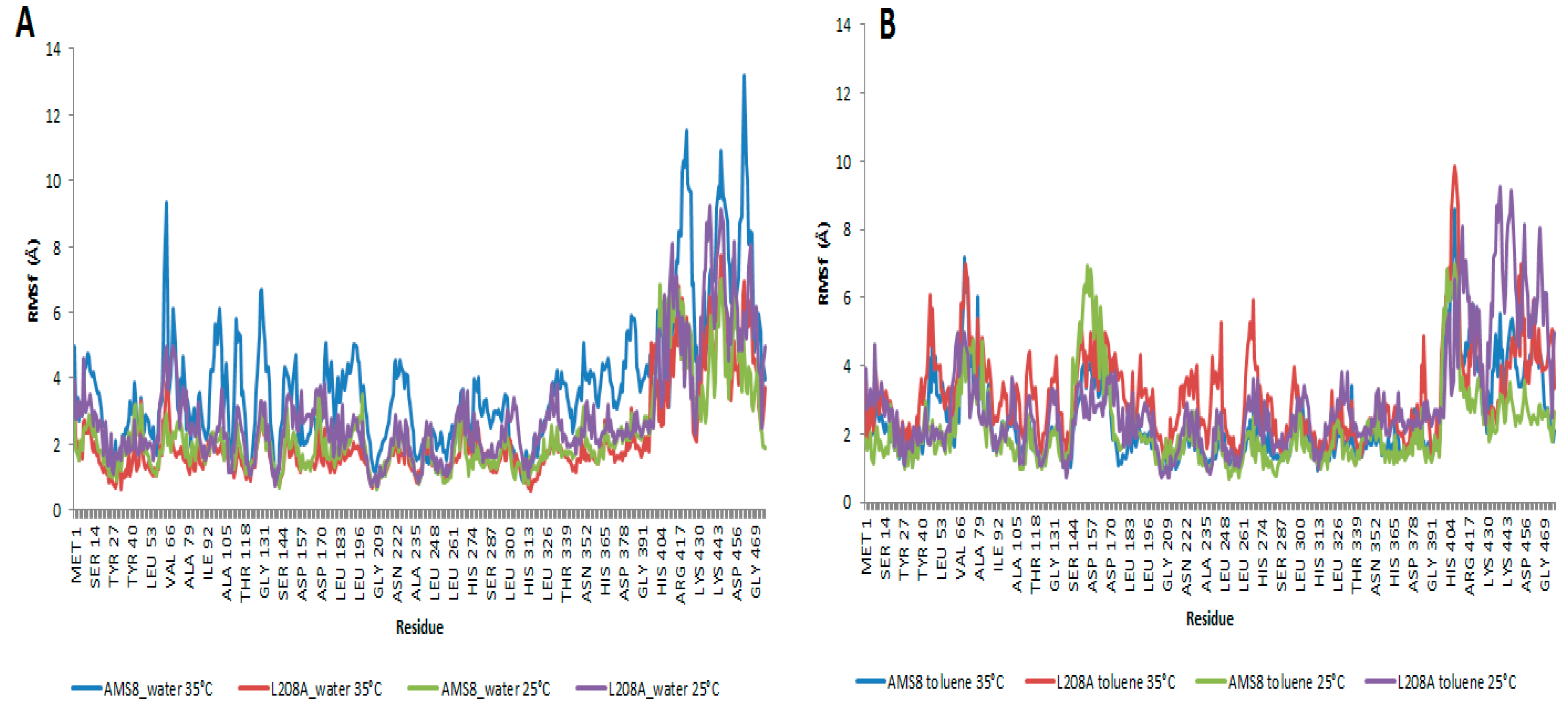
Root-mean-square fluctuations (RMSf) of the Cα atoms from residues 1–476 were calculated for the two simulation parameters. In water, WT AMS8 lipase showed a larger fluctuation starting at residues Ser-60 to Gly-65 when simulated at 35 °C compared to a similar simulation running at 25 °C. The increased motion in the simulation was mainly due to the polar properties of the affected amino acids which have been stimulated by the vibration of water molecules promoted by the increase in temperature. These regions did not embody any essential core structure such as lid region that was homology modelled and verified for its position at residues Ala-51 to Leu-57 and Glu-148 to Gly-167 which was similar to that of
Pseudomonas sp. MIS38, PML [
14]. The fluctuations would not probably change any conformation of the protein, but both N- and C-terminal domains were disturbed by the large amplitude motions caused by the temperature change. As for L208A lipase, similar residues were not perturbed by the change of temperature as observed in
Figure 4A. This caused the protein to remain stable in water. In toluene, the increased motions mainly come from three sites; the first site was lid 1 (residue 51–57), second site was Ser-60 to Gly-65 (as observed in water simulation) and the third one is lid 2 (Glu-148 to Gly-167) as shown in
Figure 4B. Key difference could be observed at the first and third sites where toluene which has been earlier found to promote lid 2 activation was also found to concurrently stimulate lid 1 opening. Apparently it hits the opening of lid 1 at temperature 35 °C as the change affects coordination of related amino acids. It was discovered at 35 °C that both WT AMS8 and L208A lipases could achieve double-lid movements in unison. For that, it is predicted that a complete interfacial activation occurred through the opening of shorter-range lid 1 and longer-range lid 2 at higher temperature can allow the access of a broader range of substrates which makes the enzyme readily receptive for various biochemical industry applications, especially those that involve toluene. After all, the water-oil interface concept may not seem to be the major requirement in enabling the movement of the lids and providing access to the catalytic pocket of the WT AMS8 lipase [
15].
Following the current interpretation, the superimposed of L208A lipase at 0 and 20 ns was structurally compared and from here, the three fluctuated sites or sequences were identified as shown in
Figure S1A. The three aforementioned sites were observed as the most flexible region as shown in the protein structure clustal alignment (
Figure S1B). Most importantly, all three catalytic residues Ser-207 (positions 207–214), Asp-255 (254–255) and His-313 (314–315) positions have completely changed and the enzyme experienced losing the native structure with the substitution of Ala-208. Thus, it was also predicted that mutant Ala-208 could hugely affects the catalytic rate via modification of substrate binding sites.
The conformational dynamics of wild type AMS8 and mutant L208A lipases showed that minimal reorganization and flexibility of lid regions was needed when the temperature was set 10 °C higher. Furthermore, a slight increase in temperature made the active sites of L208A lipase less elastic in the presence of toluene and water (supported by MD). As for wild type AMS8 lipase in water, no difference in terms of folding state and rigidity was observed when the temperature was adjusted, indicating that AMS8 lipase remained stable within the given temperature range. Many studies have inferred that the conformational flexibility of a cold-active protein is a direct consequence of conformational stabilization, where protein stability is associated with a decrease in conformation flexibility and a reduction in enzyme activity [
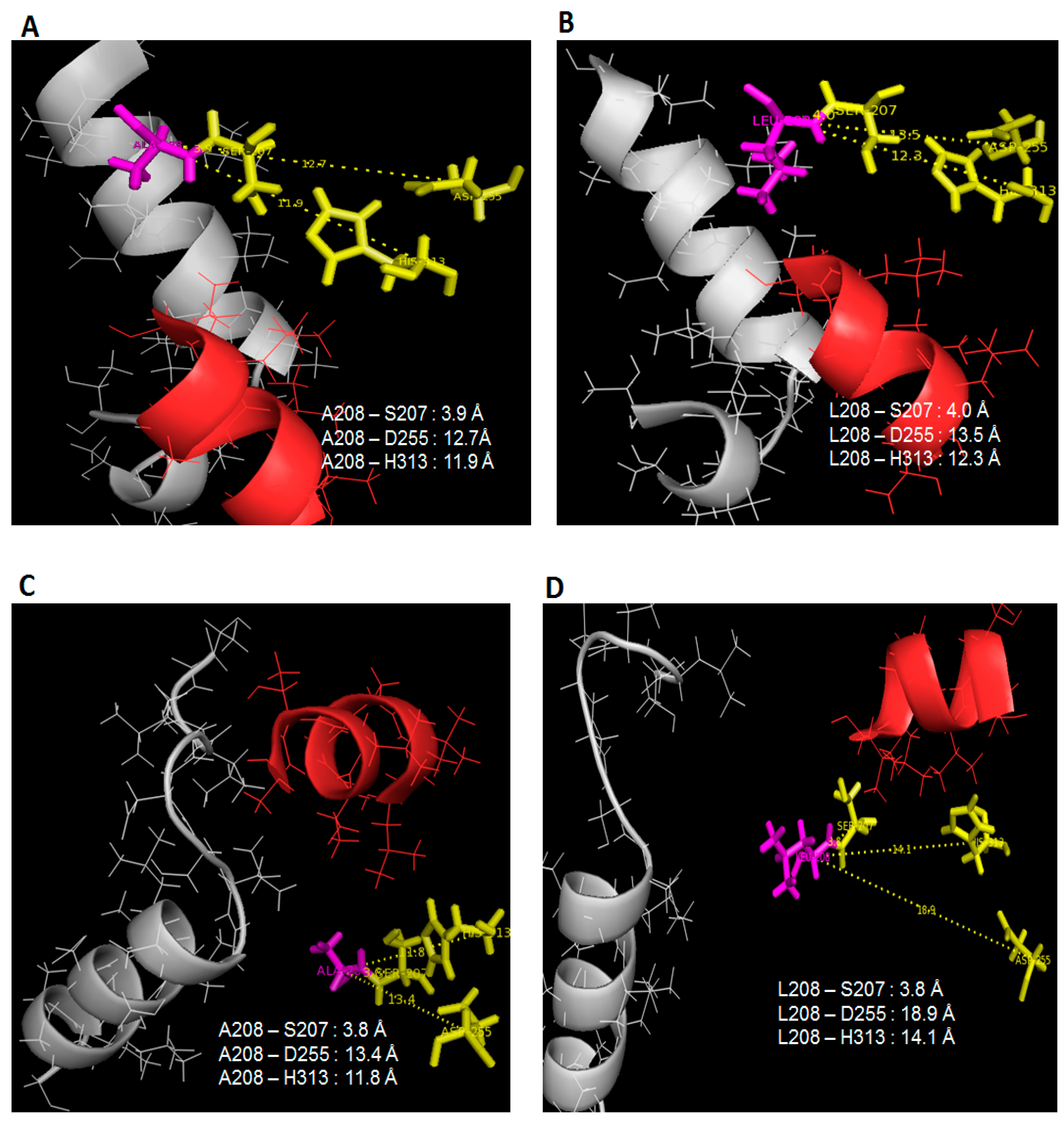
16], but in this scenario, it is rare to see such improvements in protein stability and flexibility (only at lid 1 area) almost at the same time which coincidentally was instigated by temperature changes. This finding relates to the restructuring of active sites position which alter the specific-substrate binding abilities. Following the lack of fluctuations surrounding the lid 2 region in 25 °C simulations of L208A lipase, it was found that the space that separates the nucleophilic serine from the rest of the catalytic residues has been reduced significantly, as shown in
Figure 5A,C. Once Leu-208 was changed to Ala-208, the distances between Leu-208 to Asp-255 and His-313 were reduced by 0.8 Å and 0.4 Å, respectively, in water (see
Figure 5A,B), while in toluene, the distance differences between these triads were much greater because lid 2 was opened in WT AMS8 lipase but closed in L208A lipase as a result of Ala-208 substitution which demonstrates a distance difference of 5.5 Å and 2.3 Å to Asp-255 and His-313, respectively (
Figure 6C,D). Even though the geometry of the catalytic sites (includes catalytic triad and the nucleophilic elbow motifs) is highly conserved in mutant L208A, the size and shape of the binding site differ considerably. Wild-type AMS8 lipase binds fatty acids by shoving the substrate into a long stretchy funnel-shaped binding site with contribution by hydrogen-bonds formed between Ser-238 and Gly-210 (
Figure S2A). However, in the absence of hydrogen bonding between Ser-238 and Gly-210 in the L208A mutant, the binding-site of this fatty acid was blocked due to the collapse of this tunnel (
Figure S2B). Most lipases are classified according to the binding of fatty acid moieties (from esters) into three classes, where lipases could form a hydrophobic, crevice-like binding site located near the protein surface such as the lipases from
Rhizomucor and
Rhizopus, or lipases with a funnel-like binding site, such as the lipases from
Candida antarctica, and
Pseudomonas or finally lipases with a tunnel-like scissile fatty acid binding site such as the lipases from
Candida rugosa and
Geotrichum candidum [
17]. The molecular basis of the fatty acid binding site and the role of Leu-208 in chain length specificity will be discussed later by observing the ethyl hexanoate docking analysis and kinetic profiles of various
p-nitrophenol substrates, with and without toluene (K
M and K
cat).
The temperature dependence of catalytic activity in lipase has been widely reported by researchers, with most reporting that site-directed mutagenesis targeting catalytic residues would disfavour nothing but the catalytic efficiencies. The advantage of this technique would be to improve the structure stabilization and a shift in temperature dependence [
18,
19]. Cold-active lipases were originally adapted to thrive and produce well in cold environments but due to the limitations cause by the thermolability properties, this hindered the enzyme from achieving the proper balance between stability, activity and conformational flexibility. It is now clear that AMS8 lipase is exceptionally stable and active in the temperature range between 20–25 °C as indicated by previous temperature characterization profiles [
14], but the enzyme shows that either temperature or solvent itself could initiate a significant effect on the structure stability or flexibility until a single point mutation was introduced [
15,
20].
As a consequence of having a mutated residue Ala-208, the catalytic residues of this lipase became slightly closer in distance when simulated in water and even more closer in solvent. This happened within the optimal temperature range required by AMS8 lipase for catalytic function (20–30 °C). As the temperature increased, the changes in the distance of catalytic triads could be affected by the gradual loss of proper folding and a decline in structure flexibility of this cold-active lipase. Flexibility is crucial for activity at low temperature and such alterations or substitutions made on the active site and its neighbor residues could potentially lead to substrate binding obscurity and inefficient catalysis at low temperatures. Thus it is imperative to keep the motion unrestricted [
5].
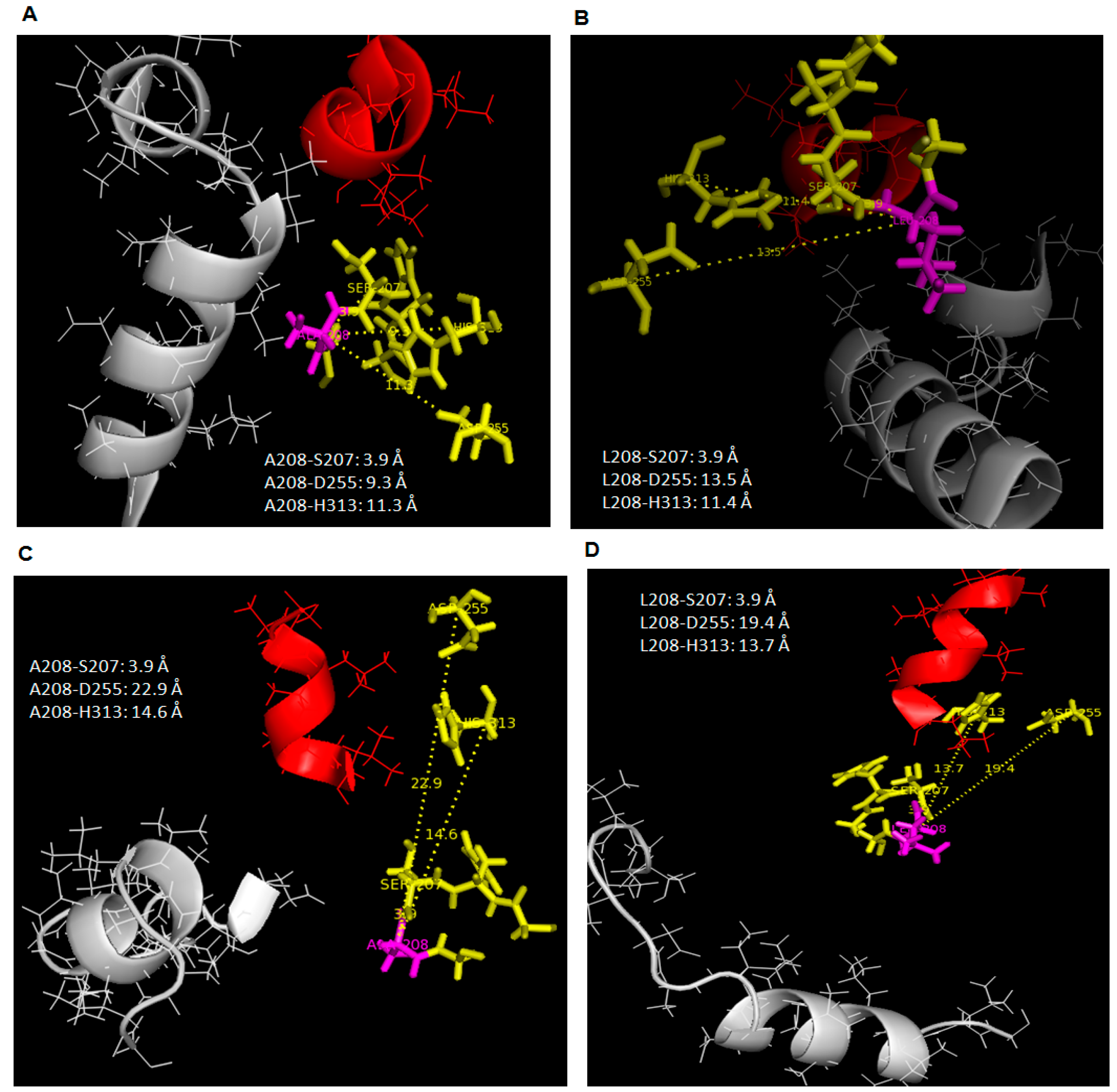
At 35 °C, all trajectories at 20 ns showed that the distance between Ala-208 to catalytic Asp-255 and His-313 had become closer than it has been (Leu-208) initially at 25 °C when simulated in water. This effect was similar to WT AMS8 lipase simulated in water (
Figure 6A,B). As a result, the catalytic binding sites will become more rigid at high temperature. Hence, this results showed an agreement with the stability (RMSd) gained by both wild-type and mutated lipases when simulated at higher temperature in the presence of water. However, the catalytic residues (Asp-255 and His-313) of both AMS8 and L208A lipases were found to drift further away from the position reported in toluene at 25 °C (
Figure 6C,D).
2.2. Electrostatic Potential and Hydrophobic Interaction Distribution of L208A and WT in Toluene
Differences in electrostatic potentials in and around the active site of psychrophilic enzymes appear to be a crucial indicator for activity at low temperatures. Electrostatic surface potentials generated by charged and polar groups are an essential component of the catalytic mechanism at various stages: as the potential extends out into the medium, a substrate can be oriented and attracted before any contact between enzyme and substrate occurs [
21]. As for hydrophobic effects, they are an important factor that influences protein folding and solubility. The solubility of non-polar compounds in water is small due to the large decrease in entropy associated with the formation of clathrate hydrate caging surrounded the nonpolar residues [
22,
23]. It is through the increase in entropy or collapse of the cage-like structure that will bring to the formation of E-S complex at the active site function as a driving force in substrate binding. This effect is called hydrophobic interactions and the higher the contribution of hydrophobic effects in a binding process, the higher will the organic solvent effect turn out to be as reflected by the changes of K
cat [
24]. Thus, the changes in hydrophobic and electrostatic energy by toluene would be reflected based on the K
M and K
cat of both L208A and WT lipases. At 25 °C, it was found that electrostatic energy (kJ/mol) has a non-linear correlation with hydrophobicity (via fraction of matching hydrophobic surface) as indicated in
Table S1. Both L208A and WT lipases were shown to have a lower hydrophobic surface match when reacted in toluene as compared to water at 25 °C. This means the hydrophobicity interaction between the lipase and the substrate, ethyl hexanoate, is not improved by mutating Leu-208 to alanine, partly due to the absence of a hydrophobic side chain. The turnover number, K
cat and substrate affinity, K
M of L208A lipase that utilizes
p-nitrophenol caprylate (C-8) declined progressively in the presence of toluene, but increased in water. The decline of both K
M and K
cat of mutant L208A could be attributed to the preservation and growing numbers of hydrophilic surface which greatly opposing the polarity of toluene. With high electrostatic energy, the hydrogen-bond formed between enzyme and substrate increases. Thus, this condition promotes the distribution of hydrophilic contacts developed between the substrate and enzyme in the presence of toluene.
2.3. Substrate/Product-Enzyme Binding
Part of the rate limiting step in lipase activity is the formation of a tetrahedral intermediate stimulated by the nucleophilic attack of the catalytic serine [
25]. A tetrahedral intermediate (or some called it oxyanion hole) forms in the lipase to drive the ester hydrolysis catalysis. It is the formation or collapse of the TI that determines the substrate selectivity [
26]. The binding pockets of lipase provide a prearranged environment to specifically stabilize this intermediate by hydrogen-bonding. Therefore, a predictive model for WT AMS8 and mutant L208A lipase that rigidly docks its substrate has to take into account whether this archetype reflects the interaction of enzyme in stabilizing the transition state which causes the anionic carbonyl oxygen to move deeper into the active site. Esterification may take place, once a tetrahedral enzyme-substrate (ES) intermediate is stabilized.
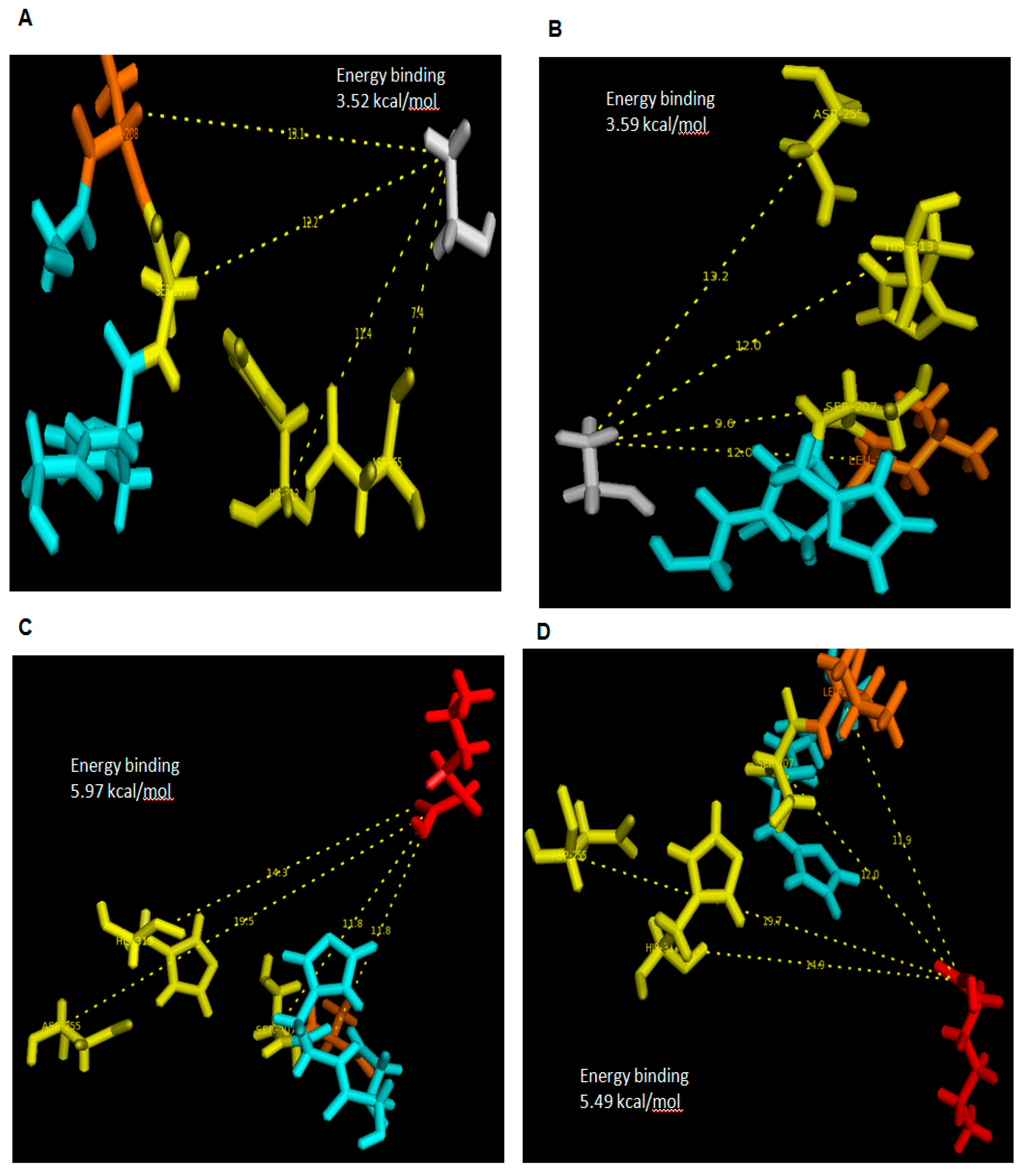
In this conventional docking, alcohol and acid substrates were covalently docked into WT and mutant L208A lipases separately at designated active site regions. After docking was completed, the binding energy involved between each substrate and lipase was determined and the value signifies the energy forces required for the formation of intermediates (enzyme + substrate). Based on the docking analyses, it was found that ethanol has a binding energy with WT AMS8 and L208A lipases of 15.02 and 14.73 kJ/mol, respectively. As for hexanoic acid, WT AMS8 and L208A were reported to have binding energies of 22.97 and 24.98 kJ/mol (
Figure 7), but when the product in the form of ethyl hexanoate were covalently docked into WT AMS8 and mutant L208A, approximate energies of 20.04 and 22.5 kJ/mol were needed for the active site binding and achieving the transition state (enzyme + product) (
Figure 8A,B). According to Koshland, the active sites of some enzymes assume a shape that is complementary to that of the transition state only after the substrate is bound. Thus, a slight increase in energy binding between mutant L208A catalytic residues and ethyl hexanoate can be associated with the lack in flexibility and steric constraints of the surrounding amino acids that form the active site [
27].
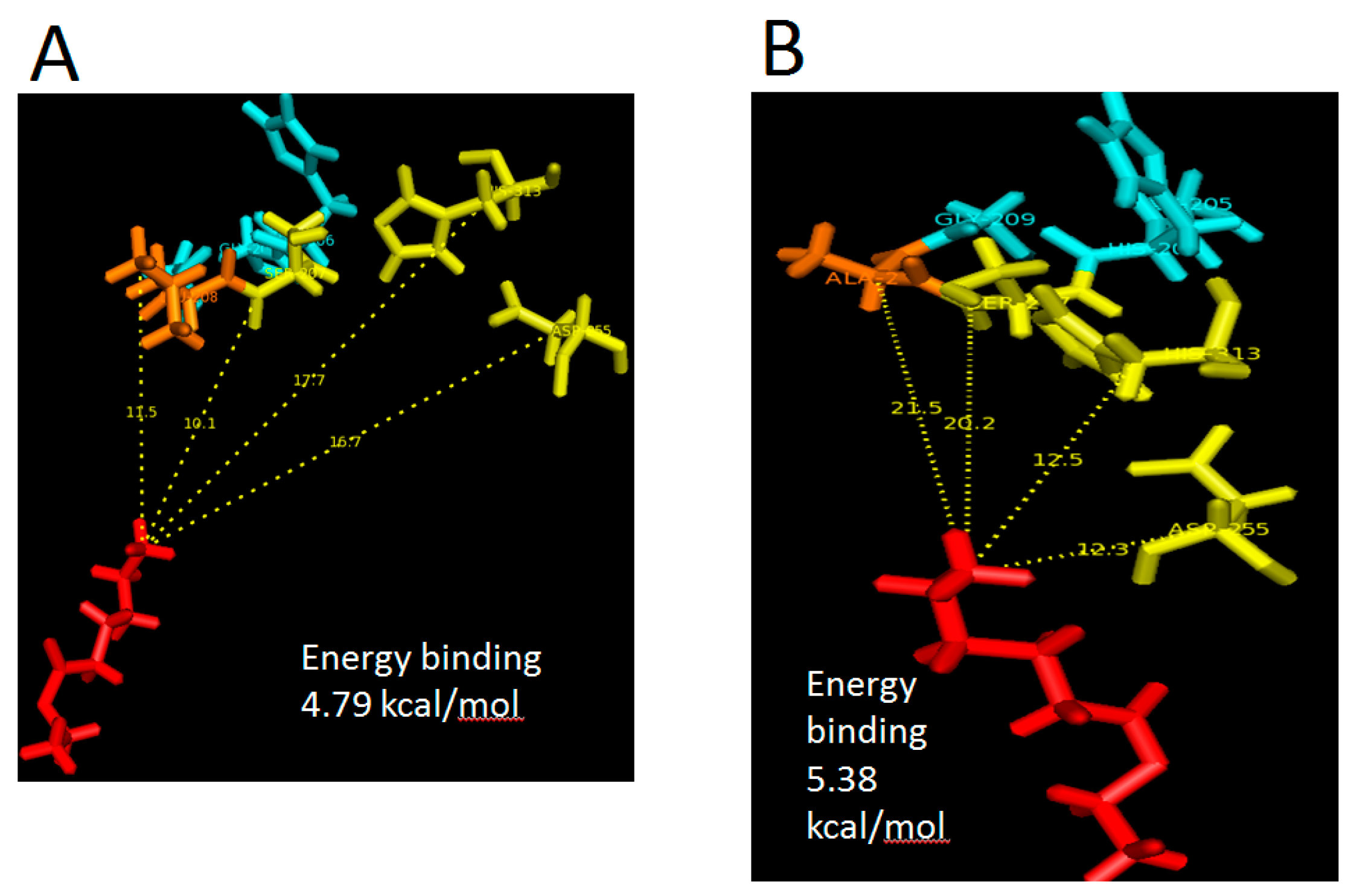
The relative binding-free energy can change prior to its binding landscape. In this case, both proteins are assumed to be rigid while the substrates are treated as a flexible molecule. In these docking analyses, it was clearly shown that Leu-208 was responsible for the binding efficiency. In the transition state, Ala-208 was placed further away from ethyl hexanoate as compared to Leu-208. The presence of a side chain in Leu-208 allows the said residue and nucleophilic serine to be placed 10 Å closer to the substrate. Meanwhile, in L208A, Asp-255 and His-313 exhibited a 5.2 and 4.4 Å closer distance with a similar substrate. Due to this reason, mutant L208A has been predicted to possess better substrate recognition due to the clustering of active site residues. However, it is also thinkable that the production or conversion rate of substrate to product will become less efficient as there will be delay for the nucleophilic serine (Ser-207) to attack the substrate due to its further distance. Mutation at substrate-binding pocket (I100F) of
Yarrowia lipolytica Lip2 lipase showed that the distance of nucleophile attack between Oγ of S162 and the carbon atom of carbonyl group in the monoester substrate, pNPC12 (I100F-pNPC12 complex) was reported to be 3.7 Å. As for WT-pNPC12 complex, a shorter distance of 3.0 Å was recorded. Similarly, the distance affects the molecular collision during reaction due to the difference in amino acid’s side-chain, decreasing the activity of I100F towards pNPC12 [
28]. However, Kumar et al. [
5] indicated that G155D substitution of lipase A from
Bacillus subtilis located at the active site assisted in a faster release of product due to the electrostatic interactions with neighboring amino acids. Therefore depending on the location of the mutation site, the conformational changes of the active site can contribute towards a variety of effects.
The strong binding energy between L208A and ethyl hexanoate also mean that this mutant lipase could spare more energy to break down the enzyme-product (EP) intermediate as a means to release the product. In this study, it is predicted that mutant L208A can cause a longer time in the formation and collapse of the intermediates (transition state) as compared to WT AMS8 lipase. Most frequently, changes that are related to active sites leads to a more accessible and/or a more flexible active site if the substitution, insertions and deletions of amino acid were made with a preference toward bulky residues [
1]. This study however, annuls such a concept in promoting active site flexibility by substituting Leu-208 with lanine, a residue adjacently located by catalytic Ser-207.
It is important that only residues which come in close contact with the substrate could tell the difference whether this substrate could form any interactions with the protein, particularly the hydrophobic ones. In general, comparisons of the best scoring docked poses for primary substrates ethanol and hexanoic acid as well as final product ester, ethyl hexanoate are shown in
Table 1. Only hexanoic acid shared a similar interaction with both proteins and the substrate apparently targeting Tyr-29, His-30, Leu-32, Ser-60, Thr-61, Ser-63, Gln-64, Gly-65, Trp-72, Ser-76, Glu-77, Arg-120 and Arg-141 which mostly located at N-terminal and these interactions were found at the surface of the two proteins. This series of residues especially positioned between 60 and 70 are highly flexible and was always moving according to B-factor analyses. These areas are rich in polar residues which could be important in forming hydrogen bond interactions between acyl group of hexanoic acid with the hydroxyl group of polar residues. In the presence of ethanol, only L208A has its catalytic site, Asp-255 interacting with it followed by subsequent residues located between Asp-255 and His-313. WT AMS8 lipase has the most contact with the ester ethyl hexanoate, particularly at the lid-2 region (Leu-166 and Gly-167) where none of this interaction occurred in mutant L208A. Thus, the substitution of residue 208 hugely affects the number of productive poses between mutant L208A and ethyl hexanoate.
In order to verify the influence of organic solvent as part of docking module, the combination of lipase/toluene/substrate system was applied by using the saved trajectory of lipase simulated in toluene to execute docking with substrates. These results would reveal the difference of substrate binding in the presence of toluene as the lipase used mimicked the one which reacted in the system. Based on the docking analysis, WT AMS8 lipase shared few contacting residues with its two substrates, and most of them were targeting residues buried inside the core of the N-domain as shown in
Table 2. This result showed that the interaction established between the substrates and buried residues of AMS8 lipase was driven by large opening left by lid 2 after simulations in toluene (
Figure S2A). Interestingly, mutant L208A surface residues located on lid 1 and lid 2 were identified as the major point of interactions that connect substrates with mutant lipase. The outcome was logical because mutant L208A provided no admission or path that could lead the two substrates straight into the catalytic area (
Figure S2B). Since lid 2 did not give ways for substrate to get inside, it was impossible to observe the binding of substrate with any residue within the catalytic surroundings. As for the end-product, ethyl hexanoate was not subjected to binding with the catalytic area in WT AMS8 lipase as it targets the residues located at N-terminal sites (situated between residues 30–78). The complex lipase docking with substrate in the presence of organic solvent has been applied in a few studies, but mostly with the objective of viewing interactions and enantioselectivity prospects in controlled systems such as microemulsions or immobilization [
29,
30].
2.4. Kinetic Profiles of L208A
To demonstrate the presence of catalytic promiscuity or substrate specificity of L208A, some relevant substrates defined by the length of carbon chain of
p-nitrophenol esters were considered for analysis. Based on the preliminary screenings, AMS8 lipase has the highest specific activity with substrates: natural oil and pNP caprylate in the presence of 25–30% (
v/
v) toluene, whereas L208A lipase displayed poor specific activity in natural oil with and without the presence of toluene. L208A lipase was found to exhibit a moderate activity in pNP-caprylate without toluene and showed highest activity with a medium-chain ester which was pNP-laurate (
Table 3). Because of these variations in substrate preference, both lipases were subjected to Michaelis-Menten kinetics to measure substrate affinity (K
M), turnover number (K
cat) and K
cat/K
M. According to the kinetic analyses, L208A demonstrated the lowest K
M and highest K
cat/K
M for the C-8 substrate (
Table 4). However, both V
Max and K
cat was found to be low, which explained the obstacle in getting a high enzymatic rate. Hence, the kinetic outcomes of mutant L208A follows in agreement with the previous protein-substrate docking analyses in associating the strength of substrate binding with energy (kJ/mol) or time (second) spent to liberate the product as a final output. In toluene, an increase of K
M value in both WT AMS8 and mutant L208A were displayed according to the substrate’s numbers of carbon-chain as it favors the smallest chain. In this case, mutant L208A has failed to promote better hydrolysis reaction in the presence of long-chain pNP substrate and toluene. For WT AMS8 lipase, the changes in K
M had significantly lower the V
max but did not result in any significant improvements to the K
cat and K
cat/K
M. The K
cat/K
M in AMS8 lipase was meagerly raised in toluene in the presence of all three pNP substrates. In contrast, mutant L208A showed a high value of K
cat/K
M with toluene only in the presence of caprylate ester.
Based on this kinetic analysis, it is interesting to learn that mutant L208A is able to hydrolyze the short pNP substrate at high K
cat/K
M (and low K
M) although facing disadvantages in the formation of a rigid active site. The idea of having a rigid active site capable of supporting higher enzyme activity in a cold-active lipase is quite contrary. Supposedly mutations that leave more space lead to a better improvement of enzyme activity [
12]. As for L208A mutation, the active sites and lid 2 regions had been reorganized or reshaped to a level of extreme rigidity which has been addressed in both molecular dynamic and docking studies. However, the mutant was still capable of making the smallest (C-8) chain substrate bind and hydrolyzed it with the highest efficiency. Both long and medium-chain substrates are thought to have difficulties in accessing the active sites due to the closing of the lid and shrinking space inside the catalytic pocket.
2.5. Esterification of Ethanol and Hexanoic Acid
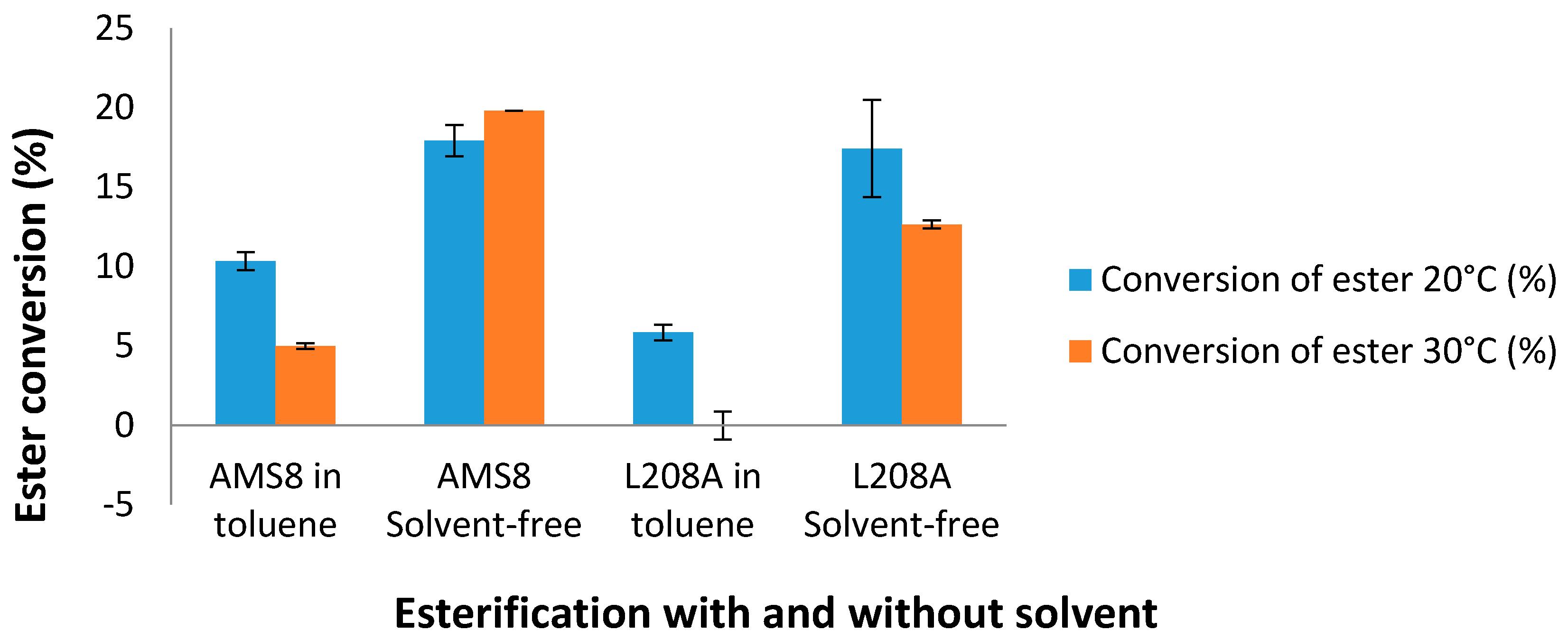
In the manufacturing industry, hexanoic acid is commercially used for the production of esters used in perfumery and in the manufacture of dyes. Therefore, L208A mutant was subjected for low-molecular weight esterification with and without the presence of toluene. Esterification of L208A lipase with the substrates ethanol and hexanoic acid at 20 °C showed that the rate of ester conversion was very much alike with AMS8 lipase in a solvent-free system as shown in
Figure 9. AMS8 lipase was found to be active over a broad temperature range of 20–30 °C [
31]. As the temperature was increased to 30 °C, the ester conversion was found to be improved by 10.5% in the solvent-free WT AMS8 lipase esterification. However, the ester conversion was reduced by 27.4% compared to the one synthesized at 20 °C in mutant L208A lipase. In toluene, WT AMS8 lipase showed a gradual decay of ester production while L208A was found not to produce esters at this temperature. Higher ester conversion in WT AMS8 lipase means that having highly flexible active site is important to facilitate rapid production of ethyl hexanoate. Thus, the smaller active site and rigidity of lid 2 from mutant L208A may not ease the effect of a rate limiting step on the reaction rate [
22]. Currently, both lipases are found not to be effective ester producers in toluene.
The replacement of Leu-208 by alanine caused a negative effect on lipase activity in the presence of toluene following the falling trend of ethyl hexanoate esterification. This outcome was anticipated as kinetic studies also showed that toluene contributes to a lower substrate binding affinity. Mutant L208A lipase activity however was found to increase by means of K
M and K
cat of pNP-caprylate hydrolysis at 25 °C and preserved ethyl hexanoate esterification at 20 °C showing that it works well in the 20–25 °C temperature range. An increase in temperature (30 °C) could cause rapid atomic movements of solvent (in toluene) and a loss of important interactions due to the shifting. A simple molecular dynamics of mutant L208A in the presence of ethyl hexanoate at 20 and 30 °C clearly exhibits a change in carbon backbone stability as shown by the RMSd result in
Figure S3A. At 30 °C, L208A experienced a major structural change after 7.5 nanoseconds of simulation preventing the mutant from having a structure amenable to ligand binding. A contrast RMSd at 20 °C, showed a flat graph with values from 2–4 Å could be an indication of an optimized structure for ligand-protein binding. The same results could be observed in the fluctuation ratio of residues in
Figure S3B where all of the structure consistency such as ligand binding, energy and interaction was highly dependent on it. Adding to the findings of L208A, the mutant also demonstrates a large radius of gyration or less compactness in structure at 30 °C (
Figure S4A). In actual fact, the compactness is a definition of a ratio of the accessible surface area of a protein to the surface area of the ideal sphere of the same volume. This may suggest that L208A has an indiscriminate region or broader sites in allowing ethyl hexanoate to bind at 30 °C. According to Lobanov et al. [
32] a compact packing of amino acid residues is known to affect both the stability and folding rate of proteins. In addition, α-proteins fold more rapidly and are less compact as compared with the β- and (α + β)-proteins, while α/β proteins have the lowest folding rate and are most compact. In this analysis, L208A demonstrates a consequent loss of α (3%) and β (1%) structure at 30 °C which had contributed towards loose packing and possibly affect its folding (
Figure S4B). Since α/β-proteins would display the lowest radius of gyration and tightest packing, a decline in α/β structures should have a major consequence in the compactness of protein structure.