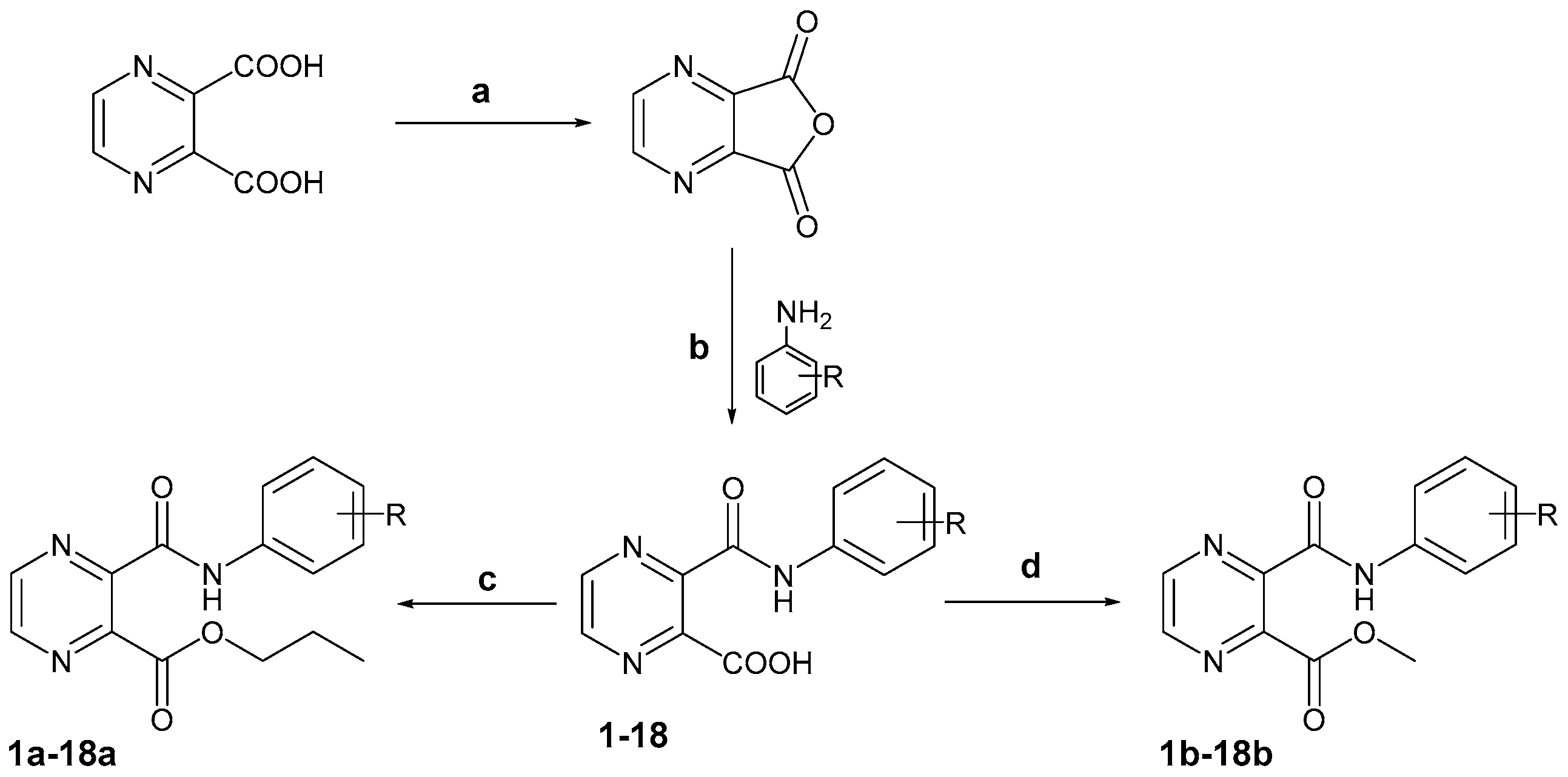
3.2.3. Analytical Data of Prepared Compounds
3-(Phenylcarbamoyl)pyrazine-2-carboxylic acid (
1) [
31]. Pale yellow solid. Yield 91%; m.p. 163.1–164.6 °C; IR (cm
−1): 3341 (N-H, CONH), 3051 (O-H, COOH), 1712 (C=O, COOH), 1682 (C=O, CONH);
1H-NMR δ 13.77 (bs, 1H, COOH), 10.75 (s, 1H, NH), 8.92–8.86 (m, 2H, pyr.), 7.80–7.75 (m, 2H, Ar), 7.40–7.34 (m, 2H, Ar), 7.16–7.11 (m, 1H, Ar);
13C-NMR δ 166.45, 162.61, 146.40, 145.85, 145.80, 144.69, 138.57, 128.98, 124.39, 120.25; Elemental analysis: calc. for C
12H
9N
3O
3 (MW 243.22): 59.26% C, 3.73% H, 17.28% N; found 59.34% C, 3.61% H, 17.24% N.
3-[(2-Hydroxyphenyl)carbamoyl]pyrazine-2-carboxylic acid (2). Yellow solid. Yield 95%; m.p. 266.4–269.3 °C; IR (cm−1): 3278 (O-H, OH), 3106 (N-H, CONH), 2854 (O-H, COOH), 1713 (C=O, COOH), 1611 (C=O, CONH); 1H-NMR δ 13.74 (bs, 1H, COOH), 10.31 (s, 1H, OH), 10.15 (s, 1H, NH), 8.93 (d, J = 2.5 Hz, 1H, pyr.), 8.90 (d, J = 2.5 Hz, 1H, pyr.), 8.24–8.20 (m, 1H, Ar), 7.03–6.93 (m, 2H, Ar), 6.88–6.83 (m, 1H, Ar); 13C-NMR δ 166.92, 160.10, 147.86, 147.12, 146.85, 144.27, 141.64, 125.81, 125.03, 119.93, 119.48, 115.13; Elemental analysis: calc. for C12H9N3O4 (MW 259.22): 55.60% C, 3.50% H, 16.21% N; found 55.77% C, 3.39% H, 16.07% N.
3-[(4-Methoxyphenyl)carbamoyl]pyrazine-2-carboxylic acid (3). Yellow solid. Yield 98%; m.p. 167.1–168.0 °C; IR (cm−1): 3330 (N-H, CONH), 3169 (O-H, COOH), 1757 (C=O, COOH), 1665 (C=O, CONH), 1243 (C-O, OCH3); 1H-NMR δ 13.74 (bs, 1H, COOH), 10.64 (s, 1H, CONH), 8.89–8.86 (m, 2H, pyr.), 7.73–7.68 (m, 2H, Ar), 6.96–6.92 (m, 2H, Ar), 3.74 (s, 3H, CH3); 13C-NMR δ 166.56, 161.97, 156.06, 146.67, 145.76, 145.47, 144.51, 131.65, 121.79, 114.08, 55.40; Elemental analysis: calc. for C13H11N3O4 (MW 273.25): 57.14% C, 4.06% H, 15.38% N; found 56.68% C, 3.93% H, 15.13% N.
3-[(2,4-Dimethoxyphenyl)carbamoyl]pyrazine-2-carboxylic acid (4). Yellow solid. Yield 85%; m.p. 199.6–200.3 °C; IR (cm−1): 3309 (N-H, CONH), 3129 (O-H, COOH), 1741 (C=O, COOH), 1667 (C=O, CONH); 1H-NMR δ 13.73 (bs, 1H, COOH), 9.95 (s, 1H, NH), 8.91 (d, J = 2.4 Hz, 1H, pyr.), 8.87 (d, J = 2.4 Hz, 1H, pyr.), 8.11 (d, J = 8.8 Hz, 1H, Ar), 6.70 (d, J = 2.6 Hz, 1H, Ar), 6.58–6.55 (m, 1H, Ar), 3.89 (s, 3H, CH3), 3.77 (s, 3H, CH3); 13C-NMR δ 166.87, 160.06, 157.18, 150.63, 147.69, 146.60, 144.22, 142.07, 121.24, 119.89, 104.50, 99.14, 56.27, 55.56; Elemental analysis: calc. for C14H13N3O5 (MW 303.27): 55.45% C, 4.32% H, 13.86% N; found 55.46% C, 4.31% H, 13.84% N.
3-[(2,5-Dimethylphenyl)carbamoyl]pyrazine-2-carboxylic acid (5). Grey solid. Yield 81%; m.p. 177.4–178.8 °C; IR (cm−1): 3339 (N-H, CONH), 2918 (O-H, COOH), 1729(C=O, COOH),1687 (C=O, CONH); 1H-NMR δ 13.75 (bs, 1H, COOH), 10.15 (s, 1H, NH), 8.91–8.87 (m, 2H, pyr.), 7.40–7.38 (m, 1H, Ar), 7.14 (d, J = 7.7 Hz, 1H, Ar), 6.99–6.95 (m, 1H, Ar), 2.28 (s, 3H, CH3), 2.22 (s, 3H, CH3); 13C-NMR δ 166.53, 162.37, 146.50, 145.90, 145.30, 144.69, 135.48, 135.36, 130.39, 129.06, 126.63, 125.43, 20.80, 17.39; Elemental analysis: calc. for C14H13N3O3 (MW 271.28): 61.99% C, 4.83% H, 15.49% N; found 62.46% C, 4.73% H, 15.53% N.
3-[(4-Ethylphenyl)carbamoyl]pyrazine-2-carboxylic acid (6). Beige solid. Yield 98%; m.p. 159.1–159.9 °C; IR (cm−1): 3314 (N-H, CONH), 2972 (O-H, COOH), 1704 (C=O, COOH), 1664 (C=O, CONH); 1H-NMR δ 10.68 (s, 1H, NH), 8.90–86 (m, 2H, pyr.), 7.71–7.65 (m, 2H, Ar), 7.23–7.16 (m, 2H, Ar), 2.58 (q, J = 7.6 Hz, 2H, CH2), 1.17 (t, J = 7.6 Hz, 3H, CH3); 13C-NMR δ 166.50, 162.32, 146.52, 145.79, 145.67, 144.60, 139.87, 136.24, 128.16, 120.32, 27.85, 15.88; Elemental analysis: calc. for C14H13N3O3 (MW 271.28): 61.99% C, 4.83% H, 15.49% N; found 61.51% C, 5.24% H, 15.08% N.
3-[(4-Fluorophenyl)carbamoyl]pyrazine-2-carboxylic acid (
7) [
27]. Grey solid. Yield 90%; m.p. 168.7–169.6 °C; IR (cm
−1): 3358 (N-H, CONH), 3069 (O-H, COOH), 1730 (C=O, COOH), 1683 (C=O, CONH);
1H-NMR δ 13.79 (bs, 1H, COOH), 10.85 (s, 1H, CONH), 8.92–8.87 (m, 2H, pyr.), 7.84–7.78 (m, 2H, Ar), 7.24–7.18 (m, 2H, Ar);
13C-NMR δ 166.46, 162.48, 158.75 (d,
J = 240.8 Hz), 146.53, 145.93, 145.51, 144.66, 134.95 (d,
J = 2.5 Hz), 122.16 (d,
J = 8.0 Hz), 115.59 (d,
J = 22.3 Hz); Elemental analysis: calc. for C
12H
8FN
3O
3 (MW 261.21): 55.18% C, 3.09% H, 16.09% N; found 54.96% C, 3.15% H, 15.85% N.
3-[(2,4-Difluorophenyl)carbamoyl]pyrazine-2-carboxylic acid (8). Grey solid. Yield 94%; m.p. 187.0–188.0 °C; IR (cm−1): 3346 (N-H, CONH), 2906 (O-H, COOH), 1702 (C=O, COOH), 1654 (C=O, CONH);1H-NMR δ 13.79 (bs, 1H, COOH), 10.54 (s, 1H, NH), 8.91 (d, 1H, J = 2.7 Hz, pyr.), 8.89 (d, 1H, J = 2.7 Hz, pyr.), 7.86–7.79 (m, 1H, Ar), 7.42–7.35 (m, 1H, Ar), 7.17–7.12 (m, 1H, Ar); 13C-NMR δ 165.4, 162.7, 159.5 (dd, J = 245.1 Hz, J = 11.4 Hz), 155.1 (dd, J = 249.9 Hz, J = 12.4 Hz), 146.4, 146.2, 144.8, 144.6, 126.7 (dd, J = 9.6 Hz, J = 2.9 Hz), 122.0 (dd, J = 12.4 Hz, J = 3.9 Hz), 111.5 (dd, J = 21.9 Hz, J = 3.8 Hz), 104.6 (dd, J = 26.6 Hz, J = 23.9 Hz); Elemental analysis: calc. for C12H7F2N3O3 (MW 279.20): 51.62% C, 2.53% H, 15.05% N; found 51.41% C, 2.05% H, 14.72% N.
3-[(4-Chlorophenyl)carbamoyl]pyrazine-2-carboxylic acid (
9) [
32]. White solid. Yield 98%; m.p. 171.1–173.2 °C; IR (cm
−1): 3331 (N-H, CONH), 2584 (O-H, COOH), 1708 (C=O, COOH), 1692 (C=O, CONH);
1H-NMR δ 13.81 (bs, 1H, COOH), 10.92 (s, 1H, CONH), 8.91–8.88 (m, 2H, pyr.), 7.84–7.80 (m, 2H, Ar), 7.45–7.41 (m, 2H, Ar);
13C-NMR δ 166.38, 162.70, 146.41, 145.97, 145.52, 144.70, 137.53, 128.90, 128.05, 121.83; Elemental analysis: calc. for C
12H
8ClN
3O
3 (MW 277.66): 51.91% C, 2.90% H, 15.13% N; found 52.24% C, 2.83% H, 15.25% N.
3-[(3,4-Dichlorophenyl)carbamoyl]pyrazine-2-carboxylic acid (10). White solid. Yield 85%; m.p. 290.0–292.1 °C; IR (cm−1): 3274 (N-H, CONH), 2898 (O-H, COOH), 1724 (C=O, COOH), 1671 (C=O, CONH); 1H-NMR δ 11.10 (s, 1H, NH), 8.92 (d, J = 2.5 Hz, 1H, pyr.), 8.90 (d, J = 2.5 Hz, 1H, pyr.), 8.16 (d, J = 2.4 Hz, 1H, Ar), 7.77–7.71 (m, 1H, Ar), 7.63 (d, J = 8.8 Hz, 1H, Ar); 13C-NMR δ 166.35, 162.94, 146.51, 146.23, 145.03, 144.76, 138.65, 131.26, 130.96, 126.00, 121.49, 120.37; Elemental analysis: calc. for C12H7Cl2N3O3 (MW 312.11): 46.18% C, 2.26% H, 13.46% N; found 46.18% C, 2.24% H, 13.32% N.
3-[(4-Bromophenyl)carbamoyl]pyrazine-2-carboxylic acid (
11) [
33]. White solid. Yield 84%; m.p. 171.1–171.9 °C; IR (cm
−1): 3312 (N-H, CONH), 2973 (O-H, COOH), 1706 (C=O, COOH), 1662 (C=O, CONH);
1H-NMR δ 10.92 (s, 1H, NH), 8.92–8.86 (m, 2H, pyr.), 7.80–7.72 (m, 2H, Ar), 7.59–7.52 (m, 2H, Ar);
13C-NMR δ 166.39, 162.75, 146.43, 145.98, 145.55, 144.71, 137.96, 131.82, 122.20, 116.16; Elemental analysis: calc. for C
12H
8BrN
3O
3 (MW 322.12): 44.75% C, 2.50% H, 13.05% N; found 44.30% C, 2.62% H, 12.55% N.
3-[(5-Fluoro-2-methylphenyl)carbamoyl]pyrazine-2-carboxylic acid (12). Beige solid. Yield 92%; m.p. 185.0–186.1 °C; IR (cm−1): 3301 (N-H, CONH), 3187 (O-H, COOH), 1746 (C=O, COOH), 1670 (C=O, CONH); 1H-NMR δ 13.83 (bs, 1H, COOH), 10.26 (s, 1H, NH), 8.92 (d, 1H, J = 2.4 Hz, pyr.), 8.90 (d, 1H, J = 2.4 Hz, pyr.), 7.51 (dd, 1H, J = 9.1 Hz J = 2.9 Hz, Ar), 7.01 (dd, 1H, J = 9.1 Hz J = 6.9 Hz, Ar), 7.00 (dt, 1H, J = 9.1 Hz J = 2.9 Hz, Ar), 2.25 (s, 3H, CH3); 13C-NMR δ 166.4, 162.7, 160.4 (d, J = 240.3 Hz), 146.3, 146.1, 145.2, 144.9, 136.9 (d, J = 10.4 Hz), 131.8 (d, J = 8.5 Hz), 127.5 (d, J = 2.9 Hz), 112.3 (d, J = 21.0 Hz), 111.0 (d, J = 24.8 Hz), 17.1; Elemental analysis: calc. for C13H10FN3O3 (MW 275.24): 56.73% C, 3.66% H, 15.27% N; found 56.84% C, 3.81% H, 14.93% N.
3-[(2-Chloro-5-methylphenyl)carbamoyl]pyrazine-2-carboxylic acid (13). Yellow solid. Yield 90%; m.p. 190.1–191.2 °C; IR (cm−1): 3342 (N-H, CONH), 2964 (O-H, COOH), 1701 (C=O, COOH), 1664 (C=O, CONH); 1H-NMR δ 13.79 (bs, 1H, COOH), 10.34 (s, 1H, NH), 8.94 (d, J = 2.5 Hz, 1H, pyr.), 8.91 (d, J = 2.5 Hz, 1H, pyr.), 7.91 (d, J = 2.0 Hz, 1H, Ar), 7.43 (d, J = 8.2 Hz, 1H, Ar), 7.09–7.05 (m, 1H, Ar), 2.33 (s, 3H, CH3); 13C-NMR δ 166.56, 161.54, 147.20, 146.73, 144.57, 142.86, 137.65, 133.83, 129.34, 127.35, 124.58, 122.67, 20.86; Elemental analysis: calc. for C13H10ClN3O3 (MW 291.69): 53.53% C, 3.46% H, 14.41% N; found 53.88% C, 3.37% H, 14.32% N.
3-[(5-Chloro-2-hydroxyphenyl)carbamoyl]pyrazine-2-carboxylic acid (14). Yellow solid. Yield 97%; m.p. 263.1–265.1 °C; IR (cm−1): 3271 (O-H, OH), 3116 (N-H, CONH), 3048 (O-H, COOH), 1709 (C=O, COOH), 1610 (C=O, CONH); 1H-NMR δ 13.78 (bs, 1H, COOH), 10.68 (s, 1H, OH), 10.16 (s, 1H, NH), 8.94 (d, J = 2.5 Hz, 1H, pyr.), 8.90 (d, J = 2.4 Hz, 1H, pyr.), 8.29 (d, J = 2.6 Hz, 1H, Ar), 7.07–7.03 (m, 1H, Ar), 6.95 (d, J = 8.7 Hz, 1H, Ar); 13C-NMR δ 166.74, 160.59, 147.63, 146.97, 146.02, 144.37, 141.58, 126.94, 124.38, 122.70, 119.30, 116.21; Elemental analysis: calc. for C12H8ClN3O4 (MW 293.66): 49.08% C, 2.75% H, 14.31% N; found 49.57% C, 2.69% H, 14.17% N.
3-[(2-Hydroxy-5-nitrophenyl)carbamoyl]pyrazine-2-carboxylic acid (15). Pale yellow solid. Yield 90%; m.p. 290.0–291.3 °C; IR (cm−1): 3566 (O-H, OH), 3363 (N-H, CONH), 2972 (O-H, COOH), 1744 (C=O, COOH), 1676 (C=O, CONH), 1545 (N-O, NO2), 1349 (N-O, NO2); 1H-NMR δ 13.73 (bs, 1H, COOH), 12.08 (bs, 1H, OH), 10.25 (s, 1H, NH), 9.16 (d, J = 2.8 Hz, 1H, Ar), 8.95 (d, J = 2.4 Hz, 1H, pyr.), 8.91 (d, J = 2.4 Hz, 1H, pyr.), 8.00–7.96 (m, 1H, Ar), 7.11 (d, J = 9.0 Hz, 1H, Ar); 13C-NMR δ 166.66, 161.15, 153.61, 147.46, 147.04, 144.49, 141.76, 139.49, 126.02, 121.49, 115.12, 114.78; Elemental analysis: calc. for C12H8N4O6 (MW 304.22): 47.38% C, 2.65% H, 18.42% N; found 46.99% C, 3.13% H, 18.15% N.
3-[(4-Nitrophenyl)carbamoyl]pyrazine-2-carboxylic acid (
16) [
34]. Yellow solid. Yield 58%; m.p. 224.5–227.2°C; IR (cm
−1): 3281 (N-H, CONH), 3065 (O-H, COOH), 1713 (C=O, COOH), 1693 (C=O, CONH), 1543 (N-O, NO
2), 1339 (N-O, NO
2);
1H-NMR δ 13.91 (bs, 1H, COOH), 11.36 (s, 1H, CONH), 8.95–8.92 (m, 2H, pyr.), 8.30–8.25 (m, 2H, Ar), 8.06–8.02 (m, 2H, Ar);
13C-NMR δ 166.18, 163.52, 146.24, 146.06, 145.61, 144.96, 144.69, 143.13, 125.12, 120.06; Elemental analysis: calc. for C
12H
8N
4O
5 (MW 288.22): 50.01% C, 2.80% H, 19.44% N; found 50.28% C, 2.82% H, 19.35% N.
3-{[3-(Trifluoromethyl)phenyl]carbamoyl}pyrazine-2-carboxylic acid (17). Pale yellow solid. Yield 97%; m.p. 176.3–177.1°C; IR (cm−1): 3356 (N-H, CONH), 3064 (O-H, COOH), 1718 (C=O, COOH), 1694 (C=O, CONH); 1H-NMR δ 11.14 (s, 1H, NH), 8.93 (d, 1H, J = 2.4 Hz, pyr.), 8.91 (d, 1H, J = 2.4 Hz, pyr.), 8.30–8.28 (m, 1H, Ar), 8.01 (d, 1H, J = 7.9 Hz, Ar), 7.62 (t, 1H, J = 7.9 Hz, Ar), 7.50 (d, 1H, J = 7.9 Hz, Ar); 13C-NMR δ 166.4, 163.1, 146.5, 146.2, 145.2, 144.8, 139.4, 130.3, 129.7 (q, J = 31.4 Hz), 124.3 (q, J = 271.8 Hz), 123.9, 120.8 (q, J = 3.8 Hz), 116.4 (q, J = 3.8 Hz); Elemental analysis: calc. for C13H8F3N3O3 (MW 311.22): 50.17% C, 2.59% H, 13.50% N; found 49.86% C, 2.75% H, 13.42% N.
3-{[4-(Trifluoromethyl)phenyl]carbamoyl}pyrazine-2-carboxylic acid (
18) [
35]. White solid. Yield 90%; m.p. 153.5–154.7 °C; IR (cm
−1): 3306, 1706 (C=O, COOH), 1669 (C=O, CONH);
1H-NMR δ 11.13 (s, 1H, NH), 8.92–8.91 (m 1H, pyr.), 8.91–8.89 (m, 1H, pyr.), 8.08–7.96 (m, 2H, Ar), 7.77–7.70 (m, 2H, Ar);
13C-NMR δ 166.4, 163.3, 146.4, 146.1, 145.7, 144.8, 142.2, 126.3 (q,
J = 3.8 Hz), 124.5 (q,
J = 270.9 Hz), 124.4 (q,
J = 31.4 Hz), 120.2; Elemental analysis: calc. for C
13H
8F
3N
3O
3 (MW 311.22): 50.17% C, 2.59% H, 13.50% N; found 50.42% C, 2.21% H, 13.84% N.
Propyl 3-(phenylcarbamoyl)pyrazine-2-carboxylate (1a). White solid. Yield 85%; m.p. 74.9–76.5 °C; IR (cm−1): 3355 (N-H, CONH), 2961 (C-H), 1734 (C=O, COO), 1686 (C=O, CONH), 1112, 1076 (C-O, COO); 1H-NMR δ 10.82 (s, 1H, CONH), 8.94 (s, 2H, pyr.), 7.80–7.77 (m, 2H, Ar), 7.407.35 (m, 2H, Ar), 7.17–7.12 (m, 1H, Ar), 4.26 (t, J = 6.6 Hz, 2H, CH2), 1.70–1.61 (m, 2H, CH2), 0.89 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.04, 162.19, 146.14, 145.68, 145.49, 145.11, 138.36, 128.94, 124.47, 120.32, 67.46, 21.45, 10.38; Elemental analysis: calc. for C15H15N3O3 (MW 285.30): 63.15% C, 5.30% H, 14.73% N; found 63.41% C, 5.40% H, 14.61% N.
Propyl 3-[(2-hydroxyphenyl)carbamoyl]pyrazine-2-carboxylate (2a). Pale yellow solid. Yield 64%; m.p. 156.8–159.6 °C; IR (cm−1): 3348 (N-H, CONH), 2924 (C-H), 1741 (C=O, COO), 1678 (C=O, CONH), 1111, 1086 (C-O, COO); 1H-NMR δ 10.33 (s, 1H, CONH), 10.16 (s, 1H, OH), 8.96 (d, J = 2.5 Hz, 1H, pyr.), 8.95 (d, J = 2.4 Hz, 1H, pyr.), 8.22–8.17 (m, 1H, Ar), 7.04–6.93 (m, 2H, Ar), 6.89–6.82 (m, 1H, Ar), 4.31 (t, J = 6.6 Hz, 2H, CH2), 1.76–1.67 (m, 2H, CH2), 0.93 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.50, 159.83, 147.16, 146.98, 146.63, 144.83, 142.09, 125.63, 125.11, 120.01, 119.45, 115.10, 67.41, 21.49, 10.41; Elemental analysis: calc. for C15H15N3O4 (MW 301.30): 59.80% C, 5.02% H, 13.95% N; found 59.83% C, 5.15% H, 13.52% N.
Propyl 3-[(4-methoxyphenyl)carbamoyl]pyrazine-2-carboxylate (3a). White solid. Yield 43%; m.p. 102.0–102.9 °C; IR (cm−1): 3284 (N-H, CONH), 2968 (C-H), 1745 (C=O, COO), 1652 (C=O, CONH), 1159, 1089 (C-O, COO); 1H-NMR δ 10.71 (s, 1H, CONH), 8.92 (s, 2H, pyr.), 7.72–7.68 (m, 2H, Ar), 6.96–6.92 (m, 2H, Ar), 4.26 (t, J = 6.6 Hz, 2H, CH2), 3.75 (s, 3H, CH3), 1.70–1.62 (m, 2H, CH2), 0.89 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.18, 161.58, 156.12, 146.06, 145.74, 145.43, 144.97, 131.44, 121.88, 114.06, 67.40, 55.37, 21.46, 10.39; Elemental analysis: calc. for C16H17N3O4 (MW 315.33): 60.94% C, 5.43% H, 13.33% N; found 60.90% C, 5.47% H, 13.21% N.
Propyl 3-[(2,4-dimethoxyphenyl)carbamoyl]pyrazine-2-carboxylate (4a). Yellow solid. Yield 75%; m.p. 116.4–118.5 °C; IR (cm−1): 3371 (N-H, CONH), 2962 (C-H), 1736 (C=O, COO), 1677 (C=O, CONH) ), 112 , 1084 (C-O, COO); 1H-NMR δ 9.97 (s, 1H, CONH), 8.76 (d, J = 2.4 Hz, 1H, pyr.), 8.68 (d, J = 2.4 Hz, 1H, pyr.), 8.43 (d, J = 8.6 Hz, 1H, Ar), 6.54–6.50 (m, 2H, Ar), 4.47 (t, J = 6.8 Hz, 2H, CH2), 3.93 (s, 3H, CH3), 3.82 (s, 3H, CH3), 1.89–1.80 (m, 2H, CH2), 1.03 (t, J = 7.5 Hz, 3H, CH3); 13C-NMR δ 166.02, 158.94, 157.01, 149.92, 148.00, 145.69, 143.06, 142.49, 120.76, 120.42, 103.79, 98.67, 68.05, 55.84, 55.51, 21.77, 10.31; Elemental analysis: calc. for C17H19N3O5 (MW 345.36): 59.12% C, 5.55% H, 12.17% N; found 58.70% C, 5.42% H, 11.87% N.
Propyl 3-[(2,5-dimethylphenyl)carbamoyl]pyrazine-2-carboxylate (5a). White solid. Yield 78%; m.p. 111.8–112.9 °C; IR (cm−1): 3354 (N-H, CONH), 2976 (C-H), 1741 (C=O, COO), 1686 (C=O, CONH), 1145, 1079 (C-O, COO); 1H-NMR δ 9.54 (s, 1H, CONH), 8.80 (d, J = 2.4 Hz, 1H, pyr.), 8.68 (d, J = 2.5 Hz, 1H, pyr.), 8.02–8.00 (m, 1H, Ar), 7.11 (d, J = 7.6 Hz, 1H, Ar), 6.93 (d, J = 7.4 Hz, 1H, Ar), 4.47 (t, J = 6.8 Hz, 2H, CH2), 2.35 (d, J = 6.1 Hz, 6H, CH3), 1.90–1.80 (m, 2H, CH2), 1.03 (t, J = 7.5 Hz, 3H, CH3); 13C-NMR δ 165.87, 159.33, 148.23, 146.08, 143.00, 142.07, 136.74, 134.76, 130.23, 126.04, 125.13, 122.31, 68.13, 21.76, 21.17, 17.16, 10.29; Elemental analysis: calc. for C17H19N3O3 (MW 313.36): 65.16% C, 6.11% H, 13.41% N; found 64.93% C, 5.91% H, 13.47% N.
Propyl 3-[(4-ethylphenyl)carbamoyl]pyrazine-2-carboxylate (6a). White solid. Yield 80%; m.p. 59.8–61.9 °C; IR (cm−1): 3332 (N-H, CONH), 2971 (C-H), 1743 (C=O, COO), 1659 (C=O, CONH), 1154, 1090 (C-O, COO); 1H-NMR δ 10.75 (s, 1H, CONH), 8.93 (s, 2H, pyr.), 7.69 (d, J = 8.1 Hz, 2H, Ar), 7.20 (d, J = 8.1 Hz, 2H, Ar), 4.26 (t, J = 6.6 Hz, 2H, CH2), 2.58 (q, J = 7.6 Hz, 2H, CH2), 1.74–1.61 (m, 2H, CH2), 1.17 (t, J = 7.6 Hz, 3H, CH3), 0.89 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.11, 161.90, 146.10, 145.62, 145.56, 145.03, 139.97, 136.04, 128.13, 120.41, 67.42, 27.83, 21.46, 15.82, 10.38; Elemental analysis: calc. for C17H19N3O3 (MW 313.36): 65.16% C, 6.11% H, 13.41% N; found 64.97% C, 6.02% H, 12.95% N.
Propyl 3-[(4-fluorophenyl)carbamoyl]pyrazine-2-carboxylate (7a). Beige solid. Yield 72%; m.p. 100.9–103.1 °C; IR (cm−1): 3351 (N-H, CONH), 2981 (C-H), 1736 (C=O, COO), 1690 (C=O, CONH), 1159, 1103 (C-O, COO); 1H-NMR δ 10.92 (s, 1H, CONH), 8.94 (s, 2H, pyr.), 7.84–7.80 (m, 2H, Ar), 7.24–7.19 (m, 2H, Ar), 4.26 (t, J = 6.5 Hz, 2H, CH2), 1.70–1.62 (m, 2H, CH2), 0.88 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.07, 162.08, 158.80 (d, J = 241.2 Hz), 146.24, 145.63, 145.37, 145.09, 134.76 (d, J = 2.5 Hz), 122.25 (d, J = 8.0 Hz), 115.57 (d, J = 22.3 Hz), 67.46, 21.46, 10.37; Elemental analysis: calc. for C15H14FN3O3 (MW 303.29): 59.40% C, 4.65% H, 13.85% N; found 59.04% C, 4.51% H, 13.59% N.
Propyl 3-[(2,4-difluorophenyl)carbamoyl]pyrazine-2-carboxylate (8a). White solid. Yield 74%; m.p. 93.3–96.1 °C; IR (cm−1): 3358 (N-H, CONH), 2976 (C-H), 1742 (C=O, COO), 1694 (C=O, CONH), 1141, 1099 (C-O, COO); 1H-NMR δ 10.61 (bs, 1H, CONH), 8.97–8.93 (m, 2H, pyr.), 7.84–7.77 (m, 1H, Ar), 7.42–7.36 (m, 1H, Ar), 7.18–7.12 (m, 1H, Ar), 4.25 (t, J = 6.6 Hz, 2H, CH2), 1.71–1.62 (m, 2H, CH2), 0.89 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.00, 162.29, 159.62 (dd, J = 244.9, 11.6 Hz), 155.18 (dd, J = 249.7, 12.7 Hz), 146.54, 145.58, 145.24, 144.32, 126.84 (dd, J = 9.9, 2.7 Hz), 121.79 (dd, J = 12.3, 3.7 Hz), 111.54 (dd, J = 22.0, 3.9 Hz), 104.61 (dd, J = 27.0, 23.8 Hz), 67.46, 21.45, 10.35.; Elemental analysis: calc. for C15H13F2N3O3 (MW 321.28): 56.08% C, 4.08% H, 13.08% N; found 55.94% C, 4.12% H, 13.21% N.
Propyl 3-[(4-chlorophenyl)carbamoyl]pyrazine-2-carboxylate (9a). White solid. Yield 65%; m.p. 121.0–122.1 °C; IR (cm−1): 3352 (N-H, CONH), 2981 (C-H), 1737 (C=O, COO), 1690 (C=O, CONH), 1146, 1104 (C-O, COO); 1H-NMR δ 10.99 (s, 1H, CONH), 8.94 (s, 2H, pyr.), 7.83 (d, J = 8.6 Hz, 2H, Ar), 7.44 (d, J = 8.5 Hz, 2H, Ar), 4.26 (t, J = 6.5 Hz, 2H, CH2), 1.65 (h, J = 7.0 Hz, 2H, CH2), 0.88 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 164.98, 162.31, 146.29, 145.50, 145.40, 145.15, 137.34, 128.89, 128.18, 121.91, 67.49, 21.45, 10.37; Elemental analysis: calc. for C15H14ClN3O3 (MW 319.75): 56.35% C, 4.41% H, 13.14% N; found 56.50% C, 4.39% H, 13.20% N.
Propyl 3-[(3,4-dichlorophenyl)carbamoyl]pyrazine-2-carboxylate (10a). White solid. Yield 34%; m.p. 101.4–102.2 °C; IR (cm−1): 3336 (N-H, CONH), 2975 (C-H), 1735 (C=O, COO), 1697 (C=O, CONH), 1148, 1082 (C-O, COO); 1H-NMR δ 11.17 (s, 1H, CONH), 8.97–8.95 (m, 2H, pyr.), 8.16 (d, J = 2.5 Hz, 1H, Ar), 7.77 (dd, J = 8.8, 2.5 Hz, 1H, Ar), 7.64 (d, J = 8.8 Hz, 1H, Ar), 4.27 (t, J = 6.5 Hz, 2H, CH2), 1.72–1.60 (m, 2H, CH2), 0.89 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 164.92, 162.54, 146.52, 145.56, 145.18, 144.91, 138.45, 131.20, 130.93, 126.11, 121.58, 120.43, 67.53, 21.45, 10.36; Elemental analysis: calc. for C15H13Cl2N3O3 (MW 354.19): 50.87% C, 3.70% H, 11.86% N; found 50.77% C, 3.71% H, 11.49% N.
Propyl 3-[(4-bromophenyl)carbamoyl]pyrazine-2-carboxylate (11a). White solid. Yield 68%; m.p. 86.6–88.6 °C; IR (cm−1): 3337 (N-H, CONH), 2966 (C-H), 1740 (C=O, COO), 1693 (C=O, CONH), 1142, 1083 (C-O, COO); 1H-NMR δ 10.99 (s, 1H, CONH), 8.94 (s, 2H, pyr.), 7.79–7.75 (m, 2H, Ar), 7.58–7.54 (m, 2H, Ar.), 4.26 (t, J = 6.5 Hz, 2H, CH2), 1.70–1.61 (m, 2H, CH2), 0.88 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 164.97, 162.33, 146.29, 145.48, 145.41, 145.15, 137.76, 131.80, 122.26, 116.30, 67.49, 21.45, 10.38; Elemental analysis: calc. for C13H14BrN3O3 (MW 364.20): 49.47% C, 3.87% H, 11.54% N; found 49.01% C, 3.75% H, 11.18% N.
Propyl 3-[(5-fluoro-2-methylphenyl)carbamoyl]pyrazine-2-carboxylate (12a). White solid. Yield 33%; m.p. 124.8–127.0 °C; IR (cm−1): 3358 (N-H, CONH), 2973 (C-H), 1739 (C=O, COO), 1690 (C=O, CONH), 1135, 1106 (C-O, COO); 1H-NMR δ 10.34 (s, 1H, CONH), 8.96 (s, 2H, pyr.), 7.53–7.49 (m, 1H, Ar), 7.33–7.28 (m, 1H, Ar), 7.03–6.98 (m, 1H, Ar), 4.28 (t, J = 6.6 Hz, 2H, CH2), 2.24 (s, 3H, CH3), 1.72–1.64 (m, 2H, CH2), 0.91 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.09, 162.07, 160.31 (d, J = 240.5 Hz), 146.48, 145.64, 145.20, 144.59, 136.76 (d, J = 10.7 Hz), 131.77 (d, J = 9.0 Hz), 127.50 (d, J = 3.2 Hz), 112.38 (d, J = 20.7 Hz), 111.04 (d, J = 24.7 Hz), 67.47, 21.49, 17.06, 10.38; Elemental analysis: calc. for C16H16FN3O3 (MW 317.32): 60.56% C, 5.08% H, 13.24% N; found 60.14% C, 5.01% H, 12.82% N.
Propyl 3-[(2-chloro-5-methylphenyl)carbamoyl]pyrazine-2-carboxylate (13a). White solid. Yield 53%; m.p. 110.9–111.4 °C; IR (cm−1): 3323 (N-H, CONH), 2974 (C-H), 1736 (C=O, COO), 1697 (C=O, CONH), 1141, 1087 (C-O, COO); 1H-NMR δ 10.21 (s, 1H, CONH), 8.82 (d, J = 2.4 Hz, 1H, pyr.), 8.73 (d, J = 2.4 Hz, 1H, pyr.), 8.42–8.40 (m, 1H, Ar), 7.32–7.26 (m, 1H, Ar), 6.94–6.90 (m, 1H, Ar), 4.49 (t, J = 6.8 Hz, 2H, CH2), 2.38 (s, 3H, CH3), 1.90–1.82 (m, 2H, CH2), 1.04 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.79, 159.54, 148.23, 146.28, 143.18, 141.68, 138.03, 133.47, 128.72, 126.10, 121.84, 120.47, 68.17, 21.78, 21.28, 10.30; Elemental analysis: calc. for C16H16ClN3O3 (MW 333.77): 57.58% C, 4.83% H, 12.59% N; found 57.72% C, 4.87% H, 12.62% N.
Propyl 3-[(5-chloro-2-hydroxyphenyl)carbamoyl]pyrazine-2-carboxylate (14a). Beige solid. Yield 43%; m.p. 196.6–199.2 °C; IR (cm−1): 3369 (N-H, CONH), 2983 (C-H), 1689, 1542, 1339 (N-O, NO2), 1156, 1116 (C-O, COO); 1H-NMR δ 10.70 (bs, 1H, OH), 10.18 (s, 1H, CONH), 8.97 (d, J = 2.4 Hz, 1H, pyr.), 8.95 (d, J = 2.5 Hz, 1H, pyr.), 8.25 (d, J = 2.7 Hz, 1H, Ar), 7.07–7.04 (m, 1H, Ar), 6.95 (d, J = 8.6 Hz, 1H, Ar), 4.32 (t, J = 6.6 Hz, 2H, CH2), 1.75–1.68 (m, 2H, CH2), 0.93 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.34, 160.33, 147.13, 146.43, 146.09, 144.94, 141.99, 126.78, 124.50, 122.65, 119.33, 116.22, 67.48, 21.50, 10.40; Elemental analysis: calc. for C15H14ClN3O4 (MW 335.74): 53.66% C, 4.20% H, 12.52% N; found 53.24% C, 3.85% H, 12.39% N.
Propyl 3-[(2-hydroxy-5-nitrophenyl)carbamoyl]pyrazine-2-carboxylate (15a). Beige solid. Yield 58%; m.p. 171.4 °C decomposition; IR (cm−1): 3324 (N-H, CONH), 3208 (O-H, OH), 2968 (C-H), 1727 (C=O, COO), 1678 (C=O, CONH), 1094, 1071 (C-O, COO); 1H-NMR δ 12.16 (bs, 1H, OH), 10.29 (s, 1H, CONH), 9.12 (d, J = 2.9 Hz, 1H, Ar), 8.99 (d, J = 2.4 Hz, 1H, pyr.), 8.96 (d, J = 2.4 Hz, 1H, pyr.), 8.01–7.97 (m, 1H, Ar), 7.11 (d, J = 9.0 Hz, 1H, Ar), 4.33 (t, J = 6.6 Hz, 2H, CH2), 1.77–1.67 (m, 2H, CH2), 0.94 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.23, 160.90, 153.75, 147.20, 146.22, 145.06, 142.17, 139.44, 125.87, 121.63, 115.21, 114.81, 67.55, 21.52, 10.41; Elemental analysis: calc. for C15H14N4O6 (MW 346.30): 52.03% C, 4.08% H, 16.18% N; found 51.60% C, 4.31% H, 16.60% N.
Propyl 3-{[3-(trifluoromethyl)phenyl]carbamoyl}pyrazine-2-carboxylate (17a). White solid. Yield 53%; m.p. 80.7–83.3 °C; IR (cm−1): 3347 (N-H, CONH), 2974 (C-H), 1745 (C=O, COO), 1697 (C=O, CONH), 1148, 1105 (C-O, COO); 1H-NMR δ 9.69 (s, 1H, CONH), 8.82 (d, J = 2.4 Hz, 1H, pyr.), 8.68 (d, J = 2.4 Hz, 1H, pyr.), 8.02 (d, J = 1.9 Hz, 1H, Ar), 7.96–7.93 (m, 1H, Ar), 7.51 (t, J = 8.0 Hz, 1H, Ar), 7.45–7.42 (m, 1H, Ar), 4.48 (t, J = 6.8 Hz, 2H, CH2), 1.90–1.82 (m, 2H, CH2), 1.04 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.68, 159.76, 148.33, 146.49, 142.98, 141.19, 137.48, 131.55 (q, J = 32.7 Hz), 129.70, 123.74 (q, J = 272.4 Hz), 122.90, 121.48 (q, J = 3.8 Hz), 120.49, 116.62 (q, J = 4.1 Hz), 68.27, 21.76, 10.29; Elemental analysis: calc. for C16H14F3N3O3 (MW 353.30): 54.39% C, 3.99% H, 11.89% N; found 53.95% C, 4.03% H, 11.65% N.
Propyl 3-{[4-(trifluoromethyl)phenyl]carbamoyl}pyrazine-2-carboxylate (18a). White solid. Yield 60%; m.p. 102.0–102.9 °C; IR (cm−1): 3308 (N-H, CONH), 2965 (C-H), 1741 (C=O, COO), 1677 (C=O, CONH), 1110, 1070 (C-O, COO); 1H-NMR δ 11.20 (s, 1H, CONH), 8.97 (s, 2H, pyr.), 8.01 (d, J = 8.4 Hz, 2H, Ar), 7.75 (d, J = 8.6 Hz, 2H, Ar), 4.27 (t, J = 6.6 Hz, 2H, CH2), 1.69–1.61 (m, 2H, CH2), 0.88 (t, J = 7.4 Hz, 3H, CH3); 13C-NMR δ 165.16, 163.12, 146.70, 145.75, 145.62, 145.56, 142.25, 126.56 (q, J = 3.8 Hz), 124.78 (q, J = 32.1 Hz), 124.72 (q, J = 271.4 Hz), 120.61, 67.82, 21.73, 10.63; Elemental analysis: calc. for C16H14F3N3O3 (MW 353.30): 54.39% C, 3.99% H, 11.89% N; found 54.06% C, 4.05% H, 11.97% N.
Methyl 3-(phenylcarbamoyl)pyrazine-2-carboxylate (1b). Pale yellow solid. Yield 71%; m.p. 81.8–82.7 °C; IR (cm−1): 3349 (N-H, CONH), 2960 (C-H), 1735 (C=O, COO), 1678 (C=O, CONH), 1142, 1087 (C-O, COO); 1H-NMR δ 10.83 (s, 1H, NH), 8.96–8.93 (m, 2H, pyr.), 7.81–7.76 (m, 2H, Ar), 7.40–7.35 (m, 2H, Ar), 7.18–7.12 (m, 1H, Ar), 3.89 (s, 3H, CH3); 13C-NMR δ 165.58, 161.96, 146.65, 146.24, 145.52, 145.15, 138.24, 128.95, 124.56, 120.52, 53.07; Elemental analysis: calc. for C13H11N3O3 (MW 257.25): 60.70% C, 4.31% H, 16.33% N; found 60.35% C, 4.55% H, 16.79% N.
Methyl 3-[(2,4-dimethoxyphenyl)carbamoyl]pyrazine-2-carboxylate (4b). Yellow solid. Yield 59%; m.p. 192.5–194.2 °C; IR (cm−1): 3367 (N-H, CONH), 2965 (C-H), 1740 (C=O, COO), 1676 (C=O, CONH), 1124, 1084 (C-O, COO); 1H-NMR δ 9.99 (s, 1H, NH), 8.95 (d, J = 2.4 Hz, 1H, pyr.), 8.94 (d, J = 2.4 Hz, 1H, pyr.), 8.06 (d, J = 8.8 Hz, 1H, Ar), 6.71 (d, J = 2.6 Hz, 1H, Ar), 6.59–6.55 (m, 1H, Ar), 3.90 (d, J = 6.9 Hz, 6H, CH 3), 3.77 (s, 3H, CH 3); 13C-NMR δ 165.88, 159.72, 157.33, 150.75, 146.81, 146.26, 144.91, 142.39, 121.52, 119.60, 104.56, 99.12, 56.27, 55.56, 53.00; Elemental analysis: calc. for C15H15N3O5 (MW 317.30): 56.78% C, 4.77% H, 13.24% N; found 56.83% C, 4.56% H, 13.44% N.
Methyl 3-[(2,5-dimethylphenyl)carbamoyl]pyrazine-2-carboxylate (5b). Pale yellow solid. Yield 67%; m.p. 132.1–134.6 °C; IR (cm−1): 3379 (N-H, CONH), 2953 (C-H), 1735 (C=O, COO), 1683 (C=O, CONH), 1121, 1082 (C-O, COO); 1H-NMR δ 10.26 (s, 1H, NH), 8.96–8.93 (m, 2H, pyr.), 7.38–7.35 (m, 1H, Ar), 7.15 (d, J = 7.7 Hz, 1H, Ar), 7.01–6.96 (m, 1H, Ar), 3.88 (s, 3H, CH3), 2.29 (s, 3H, CH3), 2.21 (s, 3H, CH3); 13C-NMR δ 165.65, 161.75, 146.33, 145.63, 145.19, 144.69, 135.44, 135.29, 130.39, 129.19, 126.81, 125.52, 53.02, 20.77, 17.34; Elemental analysis: calc. for C15H15N3O3 (MW 285.30): 63.15% C, 5.30% H, 14.73% N; found 62.71% C, 5.49% H, 14.92% N.
Methyl 3-[(4-ethylphenyl)carbamoyl]pyrazine-2-carboxylate (6b). Pale brown solid. Yield 58%; m.p. 89.1–90.7 °C; IR (cm−1): 3356 (N-H, CONH), 2962 (C-H), 1737 (C=O, COO), 1686 (C=O, CONH), 1147, 1083 (C-O, COO); 1H-NMR δ 10.76 (s, 1H, NH), 8.95–8.92 (m, 2H, pyr.), 7.72–7.67 (m, 2H, Ar), 7.23–7.18 (m, 2H, Ar), 3.88 (s, 3H, CH3), 2.58 (q, J = 7.6 Hz, 2H, CH2), 1.19–1.14 (m, 3H, CH3); 13C-NMR δ 165.63, 161.69, 146.19, 145.62, 145.12, 145.08, 140.07, 135.92, 128.14, 120.59, 53.04, 27.84, 15.84; Elemental analysis: calc. for C15H15N3O3 (MW 285.30): 63.15% C, 5.30% H, 14.73% N; found 62.88% C, 5.23% H, 14.92% N.
Methyl 3-[(2,4-difluorophenyl)carbamoyl]pyrazine-2-carboxylate (8b). White solid. Yield 72%; m.p. 165.9–167.4 °C; IR (cm−1): 3348 (N-H, CONH), 2965 (C-H), 1739 (C=O, COO), 1693 (C=O, CONH), 1143, 1103, 1082 (C-O, COO); 1H-NMR δ 10.63 (bs, 1H, NH), 8.96 (s, 2H, pyr.), 7.82–7.74 (m, 1H, Ar), 7.43–7.36 (m, 1H, Ar), 7.18–7.11 (m, 1H, Ar), 3.87 (s, 3H, CH3); 13C-NMR δ 165.5, 162.2, 159.7 (dd, J = 245.1 Hz, J = 13.7 Hz), 155.4 (dd, J = 249.9 Hz, J = 13.5 Hz), 146.7, 145.5, 143.4, 144.1, 127.2 (dd, J = 9.6 Hz, J = 1.9 Hz), 121.7 (dd, J = 12.4 Hz, J = 3.9 Hz), 111.6 (dd, J = 22.0 Hz, J = 3.9 Hz), 104.7 (dd, J = 26.6 Hz, J = 23.8 Hz), 53.1; Elemental analysis: calc. for C13H9F2N3O3 (MW 293.23): 53.25% C, 3.09% H, 14.33% N; found 53.36% C, 2.76% H, 14.53% N.
Methyl 3-[(4-bromophenyl)carbamoyl]pyrazine-2-carboxylate (11b). White solid. Yield 66%; m.p. 128.5–130.4 °C; IR (cm−1): 3326 (N-H, CONH), 2956 (C-H), 1742 (C=O, COO), 1686 (C=O, CONH), 1174, 1071 (C-O, COO); 1H-NMR δ 10.99 (s, 1H, NH), 8.96–8.94 (m, 2H, pyr.), 7.80–7.76 (m, 2H, Ar), 7.58–7.55 (m, 2H, Ar), 3.88 (s, 3H, CH 3); 13C-NMR δ 165.51, 162.07, 146.64, 146.39, 145.18, 144.87, 137.64, 131.79, 122.45, 116.39, 53.10; Elemental analysis: calc. for C13H10BrN3O3 (MW 336.15): 46.45% C, 3.00% H, 12.50% N; found 46.11% C, 2.76% H, 12.19% N.
Methyl 3-[(5-fluoro-2-methylphenyl)carbamoyl]pyrazine-2-carboxylate (12b). White solid. Yield 76%; m.p. 174.2–176.9 °C; IR (cm−1): 3375 (N-H, CONH), 2955 (C-H), 1741 (C=O, COO), 1692 (C=O, CONH), 1142, 1112 (C-O, COO); 1H-NMR δ 10.36 (bs, 1H, NH), 8.97–8.95 (m, 2H, pyr.), 7.49 (dd, 1H, J = 9.7 Hz, J = 2.9 Hz, Ar), 7.33–7.28 (1H, m, Ar), 7.01 (dt, 1H, J = 9.7 Hz J = 2.9 Hz, Ar), 3.89 (s, 3H, CH3), 2.24 (s, 3H, CH3); 13C-NMR δ 165.5, 162.0, 160.3 (d, J = 240.3 Hz), 146.5, 145.3, 144.5, 144.1, 136.7 (d, J = 10.4 Hz), 131.8 (d, J = 9.4 Hz), 127.7 (d, J = 2.9 Hz), 112.5 (d, J = 21.0 Hz), 111.2 (d, J = 24.8 Hz), 53.1, 17.0; Elemental analysis: calc. for C14H12FN3O3 (MW 289.27): 58.13% C, 4.18% H, 14.53% N; found 57.74% C, 3.97% H, 14.26% N.
Methyl 3-[(2-chloro-5-methylphenyl)carbamoyl]pyrazine-2-carboxylate (13b). Pale yellow solid. Yield 74%; m.p. 166.7–168.8 °C; IR (cm−1): 3349 (N-H, CONH), 2956 (C-H), 1737 (C=O, COO), 1689 (C=O, CONH), 1144, 1088 (C-O, COO); 1H-NMR δ 10.43 (s, 1H, NH), 8.99–8.97 (m, 2H, pyr.), 7.86 (d, J = 2.0 Hz, 1H, Ar), 7.44 (d, J = 8.1 Hz, 1H, Ar), 7.12–7.05 (m, 1H, Ar), 3.91 (s, 3H, CH3), 2.33 (s, 3H, CH3); 13C-NMR δ 165.63, 161.15, 147.02, 146.00, 145.19, 142.85, 137.73, 133.66, 129.36, 127.60, 124.87, 123.02, 53.13, 20.83; Elemental analysis: calc. for C14H12ClN3O3 (MW 305.72): 55.00% C, 3.96% H, 13.75% N; found 55.43% C, 3.85% H, 13.88% N.
Methyl 3-[(5-chloro-2-hydroxyphenyl)carbamoyl]pyrazine-2-carboxylate (14b). Yellow solid. Yield 72%; m.p. 174.2–176.9 °C; IR (cm−1): 3316 (N-H, CONH), 2963 (C-H), 1735 (C=O, COO), 1681 (C=O, CONH), 1148, 1091 (C-O, COO); 1H-NMR δ 10.71 (s, 1H, OH), 10.19 (s, 1H, NH), 8.98 (d, J = 2.4 Hz, 1H, pyr.), 8.96 (d, J = 2.5 Hz, 1H, pyr.), 8.25 (d, J = 2.5 Hz, 1H, Ar), 7.09–7.03 (m, 1H, Ar), 6.95 (d, 1H, Ar), 3.92 (d, J = 9.0 Hz, 3H, CH3); 13C-NMR δ 165.80, 160.17, 147.20, 146.66, 146.10, 145.02, 141.77, 126.72, 124.55, 122.69, 119.34, 116.23, 53.13; Elemental analysis: calc. for C13H10ClN3O4 (MW 307.69): 50.75% C, 3.28% H, 13.66% N; found 50.39% C, 3.19% H, 13.72% N.
Methyl 3-[(2-hydroxy-5-nitrophenyl)carbamoyl]pyrazine-2-carboxylate (15b). Pale yellow solid. Yield 60%; m.p. 226.4–228.6 °C; IR (cm−1): 3317 (N-H, CONH), 2969 (C-H), 1732 (C=O, COO), 1683 (C=O, CONH), 1267, 1095 (C-O, COO); 1H-NMR δ 12.10 (bs, 1H, OH), 10.29 (s, 1H, NH), 9.10 (d, J = 2.9 Hz, 1H, Ar), 8.99 (d, J = 2.4 Hz, 1H, pyr.), 8.97 (d, J = 2.4 Hz, 1H, pyr.), 8.01–7.94 (m, 1H, Ar), 7.10 (d, J = 9.0 Hz, 1H, Ar), 3.95 (s, 3H, CH3); 13C-NMR δ 165.71, 160.66, 153.69, 147.28, 146.16, 145.12, 141.83, 139.46, 125.81, 121.65, 115.15, 114.79, 53.19; Elemental analysis: calc. for C13H10N4O6 (MW 318.25): 49.06% C, 3.17% H, 17.61% N; found 48.76% C, 3.05% H, 17.52% N.
Methyl 3-{[3-(trifluoromethyl)phenyl]carbamoyl}pyrazine-2-carboxylate (17b). Yellow solid. Yield 52%; m.p. 106.3–108.5 °C; IR (cm−1): 3357 (N-H, CONH), 2919 (C-H), 1748 (C=O, COO), 1693 (C=O, CONH), 1110, 1081 (C-O, COO); 1H-NMR δ 9.71 (bs, 1H, NH), 8.82 (d, 1H, J = 2.2 Hz, pyr.), 8.69 (d, 1H, J = 2.2 Hz, pyr.), 8.08 (s, 1H, Ar), 7.93 (d, 1H, J = 8.0 Hz, Ar), 7.51 (t, 1H, J = 8.0 Hz, Ar), 7.43 (d, 1H, J = 8.0 Hz, Ar), 4.10 (s, 3H, CH3); 13C-NMR δ 166.0, 159.8, 148.0, 146.5, 145.5, 143.1, 137.4, 131.6 (q, J = 33.3 Hz), 129.7, 123.0, 123.7 (q, J = 271.9 Hz), 121.6 (q, J = 3.8 Hz), 116.7 (q, J = 3.9 Hz), 53.4.
Methyl 3-{[4-(trifluoromethyl)phenyl]carbamoyl}pyrazine-2-carboxylate (18b). White solid. Yield 50%; m.p. 146.8–148.2 °C; IR (cm−1): 3297 (N-H, CONH), 2961 (C-H), 1746 (C=O, COO), 1691 (C=O, CONH), 1112, 1063 (C-O, COO); 1H-NMR δ 9.73 (bs, 1H, NH), 8.82 (d, 1H, J = 2.2 Hz, pyr.), 8.69 (d, 1H, J = 2.2 Hz, pyr.), 7.87–7.84 (m, 2H, Ar), 7.65–7.62 (m, 2H, Ar), 4.09 (s, 3H, CH3); 13C-NMR δ 166.0, 159.8, 148.0, 146.5, 145.5, 143.1, 141.2, 126.8 (q, J = 33.8 Hz), 126.4 (q, J = 3.8 Hz), 123.9 (q, J = 270.9 Hz), 119.7, 53.4; Elemental analysis: calc. for C14H10F3N3O3 (MW 325.25): 51.70% C, 3.10% H, 12.92% N; found 51.95% C, 3.41% H, 12.83% N.