Development of Novel Nrf2/ARE Inducers Bearing Pyrazino[2,1-a]isoquinolin Scaffold with Potent In Vitro Efficacy and Enhanced Physicochemical Properties
Abstract
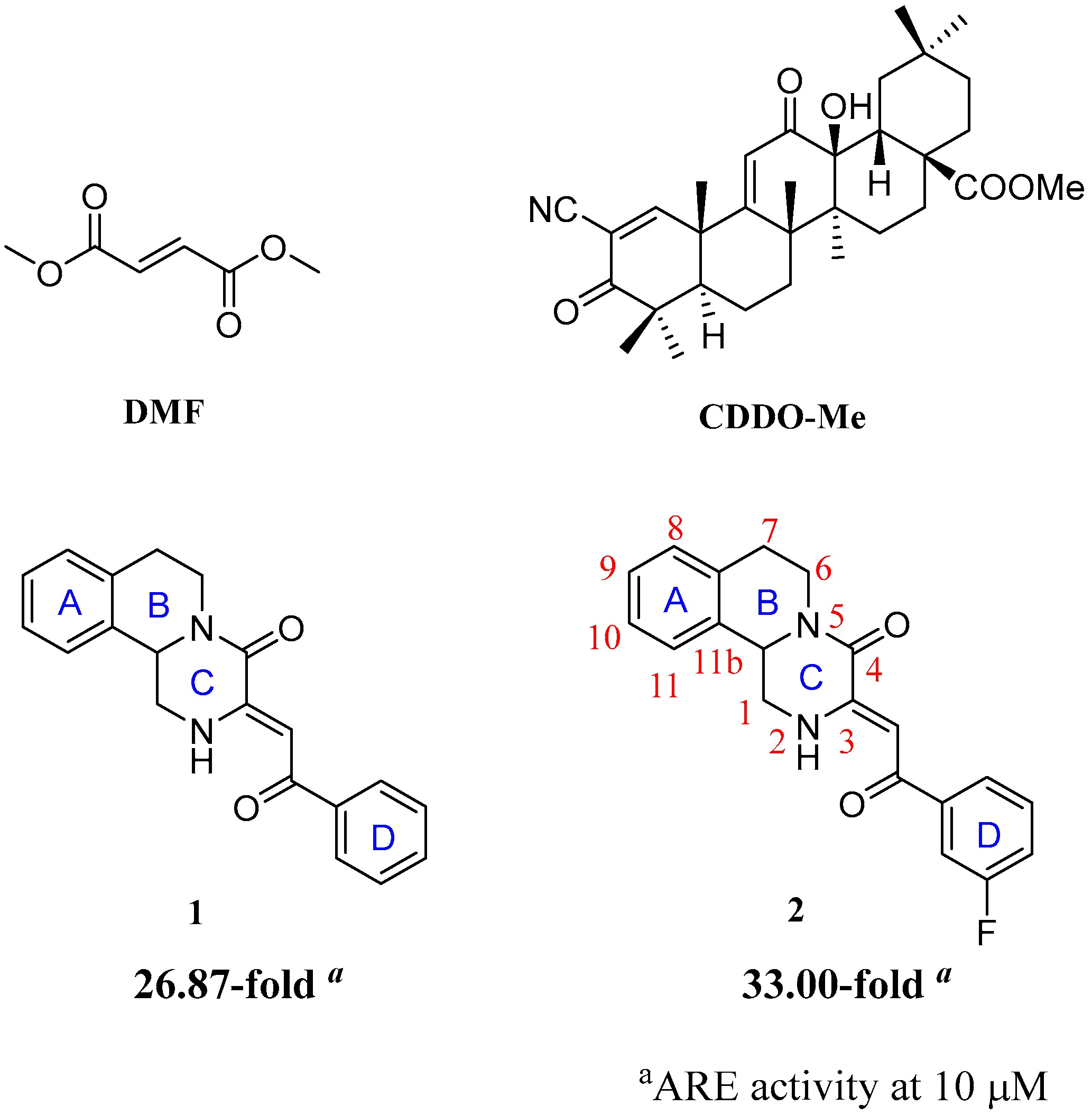
:1. Introduction
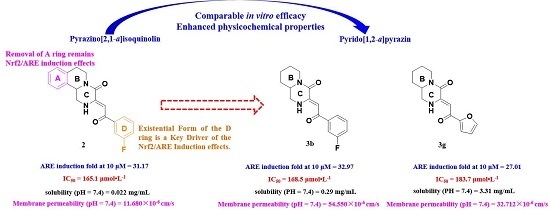
2. Results and Discussions
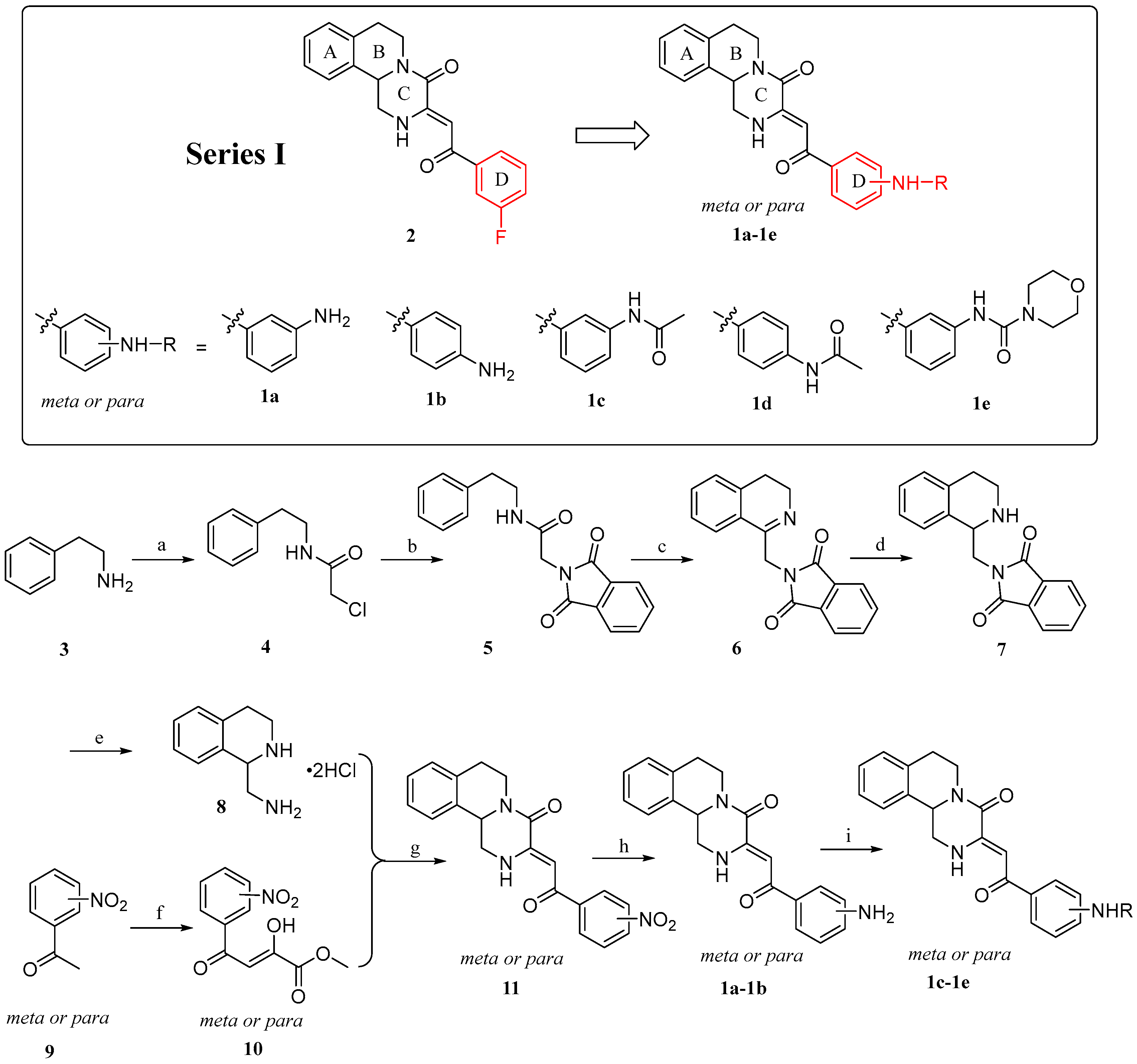
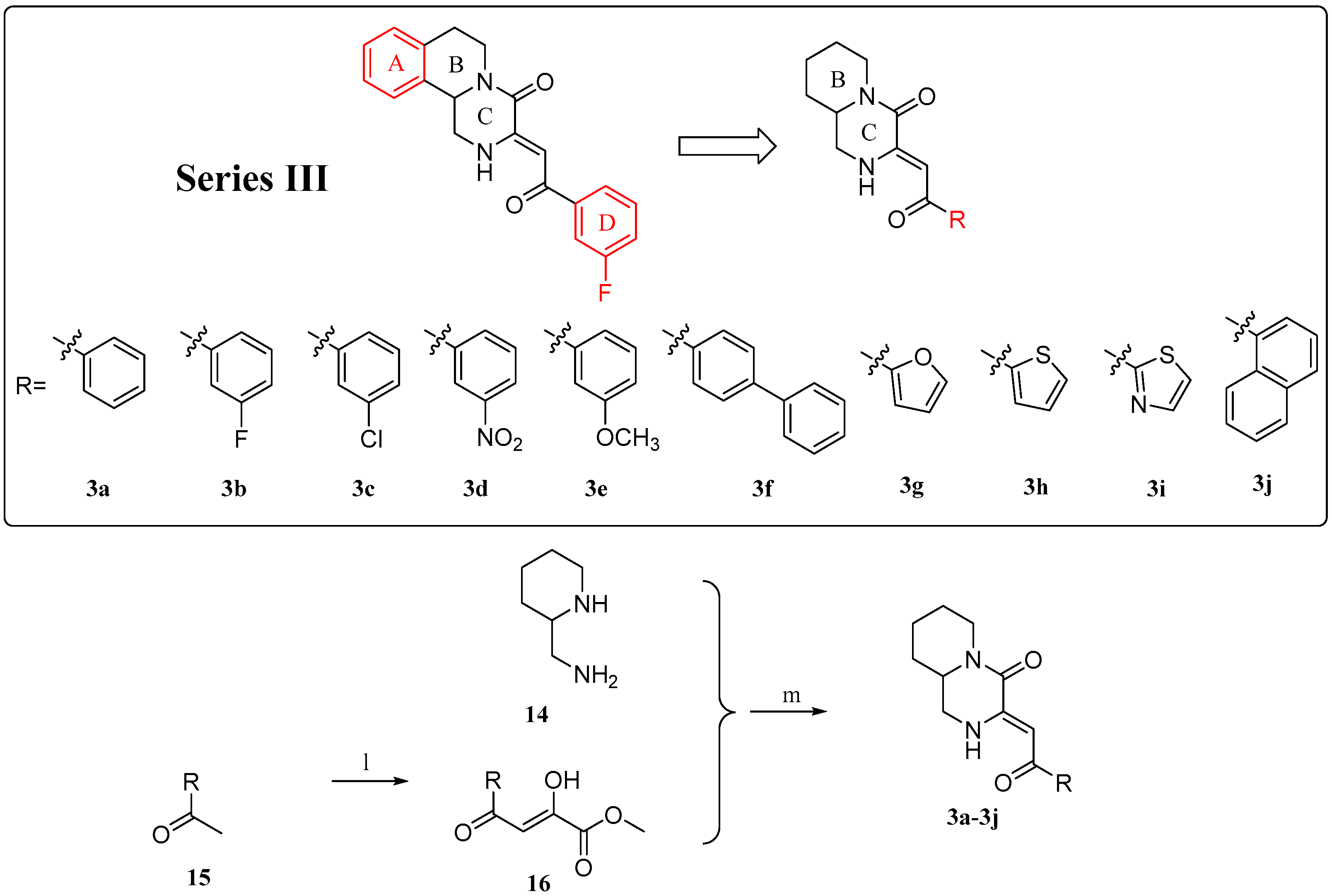
2.1. Chemistry
2.2. ARE Luciferase Reporter Assay and Cytotoxicity Evaluation
2.3. Western Blot Assay
2.4. Physicochemical Properties
3. Conclusions
4. Materials and Methods
4.1. Chemistry
4.1.1. General Chemistry
4.1.2. General Procedure for the Preparation of Compounds 1a–1e
4.1.3. General Procedure for the Preparation of Compounds 2a–2i
4.1.4. General Procedure for the Preparation of Compounds 3a–3j
4.2. Biology
4.2.1. Cell lines and Culture
4.2.2. Cell Viability Assay
4.2.3. ARE-Luciferase Activity Assay
4.2.4. Western Blot
4.3. Physicochemical Properties
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kaspar, J.W.; Niture, S.K.; Jaiswal, A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009, 47, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhou, H.-S.; You, Q.-D.; Jiang, Z. Design, Synthesis, and Initial Evaluation of Affinity-based Small-Molecule Probes for Fluorescent Visualization and Specific Detection of Keap1. J. Med. Chem. 2016, 59, 7305–7310. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yamamoto, M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2015, 88 Pt B, 93–100. [Google Scholar] [CrossRef]
- Jiang, Z.-Y.; Lu, M.-C.; You, Q.-D. Discovery and Development of Kelch-like ECH-Associated Protein 1. Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1:NRF2) Protein–Protein Interaction Inhibitors: Achievements, Challenges, and Future Directions. J. Med. Chem. 2016, 59, 10837–10858. [Google Scholar] [CrossRef] [PubMed]
- Niture, S.K.; Khatri, R.; Jaiswal, A.K. Regulation of Nrf2-an update. Free Radic. Biol. Med. 2014, 66, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Tebay, L.E.; Robertson, H.; Durant, S.T.; Vitale, S.R.; Penning, T.M.; Dinkova-Kostova, A.T.; Hayes, J.D. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015, 88 Pt B, 108–146. [Google Scholar] [CrossRef] [PubMed]
- Rojo de la Vega, M.; Dodson, M.; Chapman, E.; Zhang, D.D. NRF2-targeted therapeutics: New targets and modes of NRF2 regulation. Curr. Opin. Toxicol. 2016, 1, 62–70. [Google Scholar] [CrossRef]
- Zhuang, C.; Wu, Z.; Xing, C.; Miao, Z. Small molecules inhibiting Keap1-Nrf2 protein-protein interactions: A novel approach to activate Nrf2 function. MedChemComm 2017, 8, 286–294. [Google Scholar] [CrossRef]
- Lu, M.-C.; Ji, J.-A.; Jiang, Z.-Y.; You, Q.-D. The Keap1–Nrf2–ARE Pathway as a Potential Preventive and Therapeutic Target: An Update. Med. Res. Rev. 2016, 36, 924–963. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Kerns, J.K.; Callahan, J.F.; Moody, C.J. Keap calm, and carry on covalently. J. Med. Chem. 2013, 56, 7463–7476. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; McKercher, S.R.; Lipton, S.A. Reprint of: Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2014, 66, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Kostov, R.V.; Canning, P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch. Biochem. Biophys. 2017, 617, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Magesh, S.; Chen, Y.; Hu, L. Small Molecule Modulators of Keap1-Nrf2-ARE Pathway as Potential Preventive and Therapeutic Agents. Med. Res. Rev. 2012, 32, 687–726. [Google Scholar] [CrossRef] [PubMed]
- Xi, M.Y.; Jia, J.M.; Sun, H.P.; Sun, Z.Y.; Jiang, J.W.; Wang, Y.J.; Zhang, M.Y.; Zhu, J.F.; Xu, L.L.; Jiang, Z.Y.; et al. 3-Aroylmethylene-2,3,6,7-tetrahydro-1H-pyrazino[2,1-a]isoquinolin-4(11bH)-ones as Potent Nrf2/ARE Inducers in Human Cancer Cells and AOM-DSS Treated Mice. J. Med. Chem. 2013, 56, 7925–7938. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zheng, S.; Higgins, M.; Morra, R.P., Jr.; Mendis, A.T.; Chien, C.W.; Ojima, I.; Mierke, D.F.; Dinkova-Kostova, A.T.; Honda, T. New Monocyclic, Bicyclic, and Tricyclic Ethynylcyanodienones as Activators of the Keap1/Nrf2/ARE Pathway and Inhibitors of Inducible Nitric Oxide Synthase. J. Med. Chem. 2015, 58, 4738–4748. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.Y.; Xu, L.L.; Lu, M.C.; Chen, Z.Y.; Yuan, Z.W.; Xu, X.L.; Guo, X.K.; Zhang, X.J.; Sun, H.P.; You, Q.D. Structure-Activity and Structure-Property Relationship and Exploratory in Vivo Evaluation of the Nanomolar Keap1-Nrf2 Protein-Protein Interaction Inhibitor. J. Med. Chem. 2015, 58, 6410–6421. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.-C.; Ji, J.-A.; Jiang, Y.-L.; Chen, Z.-Y.; Yuan, Z.-W.; You, Q.-D.; Jiang, Z.-Y. An inhibitor of the Keap1-Nrf2 protein-protein interaction protects NCM460 colonic cells and alleviates experimental colitis. Sci. Rep. 2016, 6, 26585. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.-Y.; Lu, M.-C.; Xu, L.L.; Yang, T.-T.; Xi, M.-Y.; Xu, X.-L.; Guo, X.-K.; Zhang, X.-J.; You, Q.-D.; Sun, H.-P. Discovery of Potent Keap1–Nrf2 Protein–Protein Interaction Inhibitor Based on Molecular Binding Determinants Analysis. J. Med. Chem. 2014, 57, 2736–2745. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 2, 3b and 3g are available from the authors. |



| Compound | ARE Inductivity in Luciferase Reporter Assay (μM) a | IC50 (μM) | |||||
|---|---|---|---|---|---|---|---|
| 0.1 | 1 | 5 | 10 | 50 | 100 | ||
| 1a | 0.56 ± 0.28 | 1.66 ± 0.05 | 2.40 ± 1.18 | 5.64 ± 2.06 | 7.53 ± 1.28 | 7.62 ± 2.79 | >300 |
| 1b | 1.26 ± 0.28 | 1.26 ± 0.73 | 2.78 ± 0.29 | 3.45 ± 0.03 | 2.75 ± 0.68 | 2.68 ± 0.31 | >300 |
| 1c | 0.80 ± 0.19 | 0.78 ± 0.36 | 2.30 ± 0.39 | 3.05 ± 0.53 | 3.09 ± 0.09 | 3.41 ± 0.65 | >300 |
| 1d | 1.24 ± 0.23 | 1.09 ± 0.72 | 1.49 ± 0.16 | 2.94 ± 1.90 | 7.14 ± 0.34 | 10.07 ± 0.05 | >300 |
| 1e | 1.57 ± 0.05 | 1.97 ± 0.20 | 4.53 ± 0.28 | 5.48 ± 0.13 | 7.80 ± 3.94 | 34.95 ± 3.15 | 183.3 |
| 2a | 2.38 ± 0.11 | 2.28 ± 0.78 | 2.55 ± 0.31 | 3.09 ± 1.09 | 6.74 ± 0.67 | 19.60 ± 0.47 | >300 |
| 2b | 3.71 ± 1.41 | 3.57 ± 1.08 | 4.85 ± 0.82 | 6.58 ± 2.00 | 8.24 ± 0.62 | 12.90 ± 1.73 | >300 |
| 2c | 2.96 ± 0.48 | 3.04 ± 0.08 | 5.73 ± 1.44 | 13.35 ± 0.44 | 27.96 ± 1.39 | 29.01 ± 2.45 | 190.3 |
| 2d | 1.06 ± 0.09 | 3.19 ± 0.52 | 4.06 ± 0.30 | 7.38 ± 0.31 | 5.22 ± 1.28 | 5.83 ± 0.38 | 160.8 |
| 2e | 1.92 ± 0.82 | 2.17 ± 1.05 | 3.03 ± 0.68 | 3.49 ± 0.64 | 3.92 ± 0.53 | 8.31 ± 0.25 | >300 |
| 2f | 3.27 ± 1.59 | 2.10 ± 0.59 | 5.46 ± 1.83 | 8.45 ± 1.05 | 3.34 ± 1.05 | 3.07 ± 0.43 | 170.7 |
| 2g | 1.27 ± 0.52 | 2.31 ± 0.24 | 4.68 ± 0.29 | 5.84 ± 0.07 | 14.47 ± 0.36 | 9.69 ± 0.80 | >300 |
| 2h | 2.06 ± 0.98 | 3.40 ± 0.02 | 3.62 ± 0.29 | 4.59 ± 0.14 | 4.38 ± 1.94 | 4.98 ± 0.53 | 92.36 |
| 2i | 1.71 ± 0.71 | 1.88 ± 1.36 | 1.37 ± 0.19 | 1.66 ± 0.53 | 1.16 ± 0.63 | 1.42 ± 0.31 | 237.0 |
| 3a | 1.20 ± 0.49 | 4.09 ± 0.79 | 15.85 ± 2.08 | 16.38 ± 0.55 | 29.90 ± 8.49 | 40.60 ± 1.78 | >300 |
| 3b | 0.70 ± 0.35 | 5.98 ± 2.31 | 24.78 ± 2.67 | 32.97 ± 3.68 | 39.97 ± 0.69 | 4.41 ± 2.89 | 168.5 |
| 3c | 1.85 ± 0.21 | 8.41 ± 0.04 | 10.19 ± 3.44 | 11.41 ± 1.12 | 1.13 ± 1.03 | 1.33 ± 0.19 | 174.3 |
| 3d | 1.96 ± 0.88 | 3.85 ± 1.00 | 12.02 ± 2.55 | 19.52 ± 3.09 | 47.58 ± 3.54 | 8.66 ± 3.27 | 180.8 |
| 3e | 1.40 ± 1.21 | 1.82 ± 1.15 | 6.59 ± 0.77 | 9.16 ± 0.42 | 9.36 ± 3.76 | 1.44 ± 1.75 | >300 |
| 3f | 2.96 ± 0.66 | 2.21 ± 0.52 | 3.60 ± 0.42 | 1.76 ± 0.10 | 2.47 ± 0.03 | 2.69 ± 2.75 | 183.7 |
| 3g | 0.24 ± 0.15 | 1.50 ± 0.01 | 6.41 ± 3.14 | 27.01 ± 2.46 | 41.61 ± 2.60 | 79.74 ± 1.92 | >300 |
| 3h | 2.06 ± 0.22 | 2.12 ± 0.31 | 8.30 ± 2.55 | 10.52 ± 3.82 | 28.93 ± 0.46 | 1.63 ± 0.25 | >300 |
| 3i | 1.52 ± 0.87 | 4.02 ± 3.05 | 6.41 ± 1.48 | 9.28 ± 0.55 | 13.54 ± 0.80 | 16.74 ± 1.39 | >300 |
| 3j | 1.7 ± 0.19 | 2.34 ± 0.53 | 2.54 ± 0.99 | 3.52 ± 0.57 | 3.85 ± 0.34 | 1.96 ± 0.71 | 263.5 |
| 2 | 1.45 ± 0.39 | 6.20 ± 4.06 | 16.19 ± 0.30 | 31.17 ± 4.14 | 7.51 ± 1.32 | 12.16 ± 3.32 | 165.1 |
| tBHQ | 1.00 ± 0.01 | 1.80 ± 0.66 | 1.67 ± 0.56 | 1.98 ± 0.29 | 4.45 ± 1.15 | 10.79 ± 3.13 | ND |
| DMF | 1.04 ± 0.21 | 1.10 ± 0.11 | 1.40 ± 0.21 | 2.06 ± 0.25 | 3.34 ± 0.52 | 5.31 ± 0.24 | ND |
| Compound | pKa | log D, pH 7.4 | Pe (10−6 cm/s) | Solubility (mg/mL) |
|---|---|---|---|---|
| pH 7.4 | ||||
| 1e | 2.90 | 0.61 | 52.954 ± 7.290 | 0.046 |
| 2c | 3.02 | 1.22 | 43.953 ± 5.421 | 0.17 |
| 3a | 3.42 | 0.98 | 66.372 ± 5.082 | 0.53 |
| 3b | 3.10 | 1.23 | 54.550 ± 6.001 | 0.29 |
| 3g | 2.08 | −0.34 | 32.712 ± 1.922 | 3.31 |
| 2 | 2.78 | 2.38 | 11.680 ± 2.198 | 0.022 |
| Ketoprofen | - | - | 1.607 ± 0.243 | - |
| Propranolol | - | - | 64.679 ± 8.582 | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, H.; Jiao, Q.; Liu, T.; You, Q.; Jiang, Z. Development of Novel Nrf2/ARE Inducers Bearing Pyrazino[2,1-a]isoquinolin Scaffold with Potent In Vitro Efficacy and Enhanced Physicochemical Properties. Molecules 2017, 22, 1541. https://doi.org/10.3390/molecules22091541
Dai H, Jiao Q, Liu T, You Q, Jiang Z. Development of Novel Nrf2/ARE Inducers Bearing Pyrazino[2,1-a]isoquinolin Scaffold with Potent In Vitro Efficacy and Enhanced Physicochemical Properties. Molecules. 2017; 22(9):1541. https://doi.org/10.3390/molecules22091541
Chicago/Turabian StyleDai, Hongbin, Qiong Jiao, Tian Liu, Qidong You, and Zhengyu Jiang. 2017. "Development of Novel Nrf2/ARE Inducers Bearing Pyrazino[2,1-a]isoquinolin Scaffold with Potent In Vitro Efficacy and Enhanced Physicochemical Properties" Molecules 22, no. 9: 1541. https://doi.org/10.3390/molecules22091541