Characterization of Two VAO-Type Flavoprotein Oxidases from Myceliophthora thermophila
Abstract
:1. Introduction
2. Results
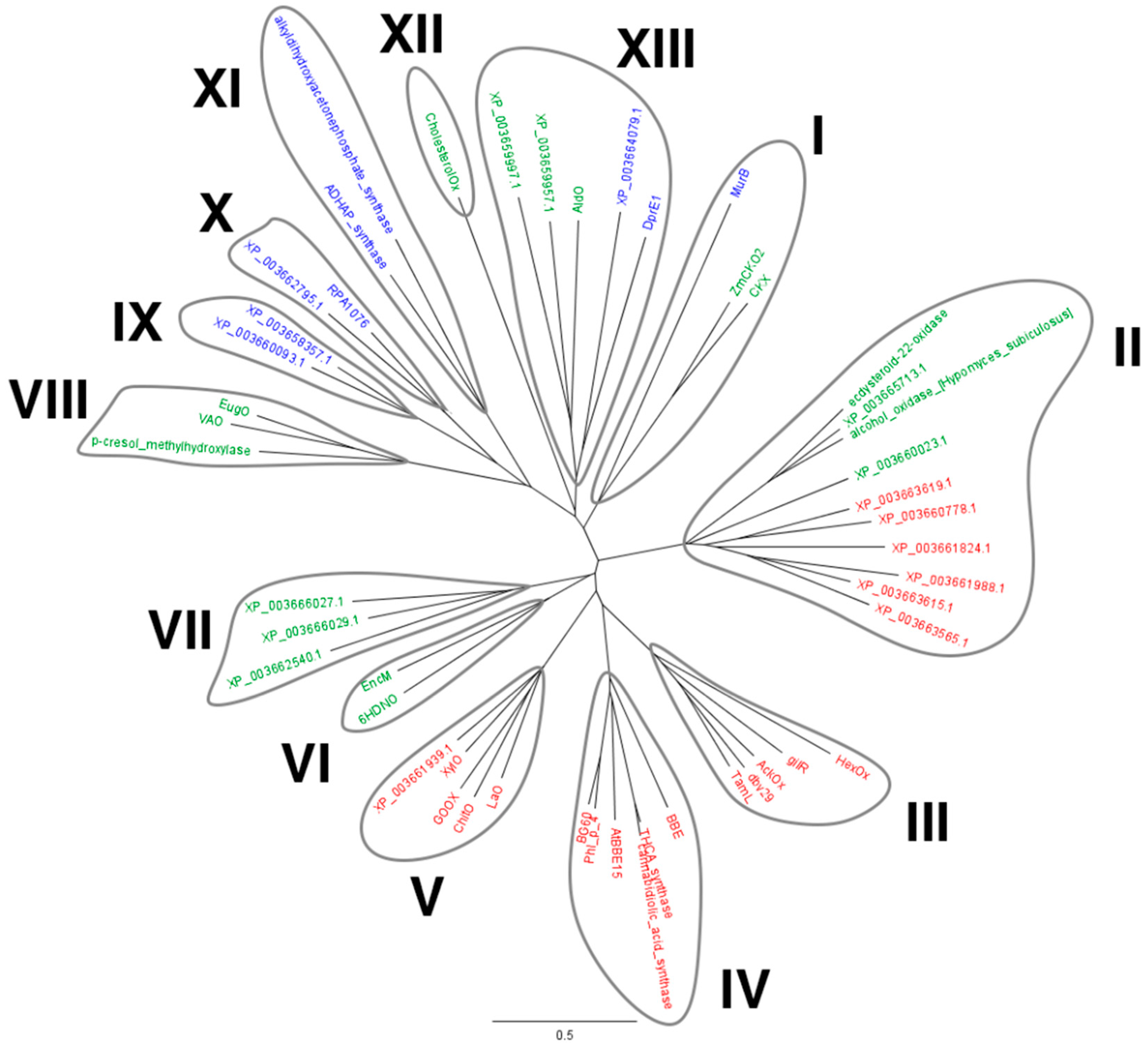
2.1. Identification of M. thermophila Genes Encoding Putative VAO-Type Flavoprotein Oxidases
2.2. Purification
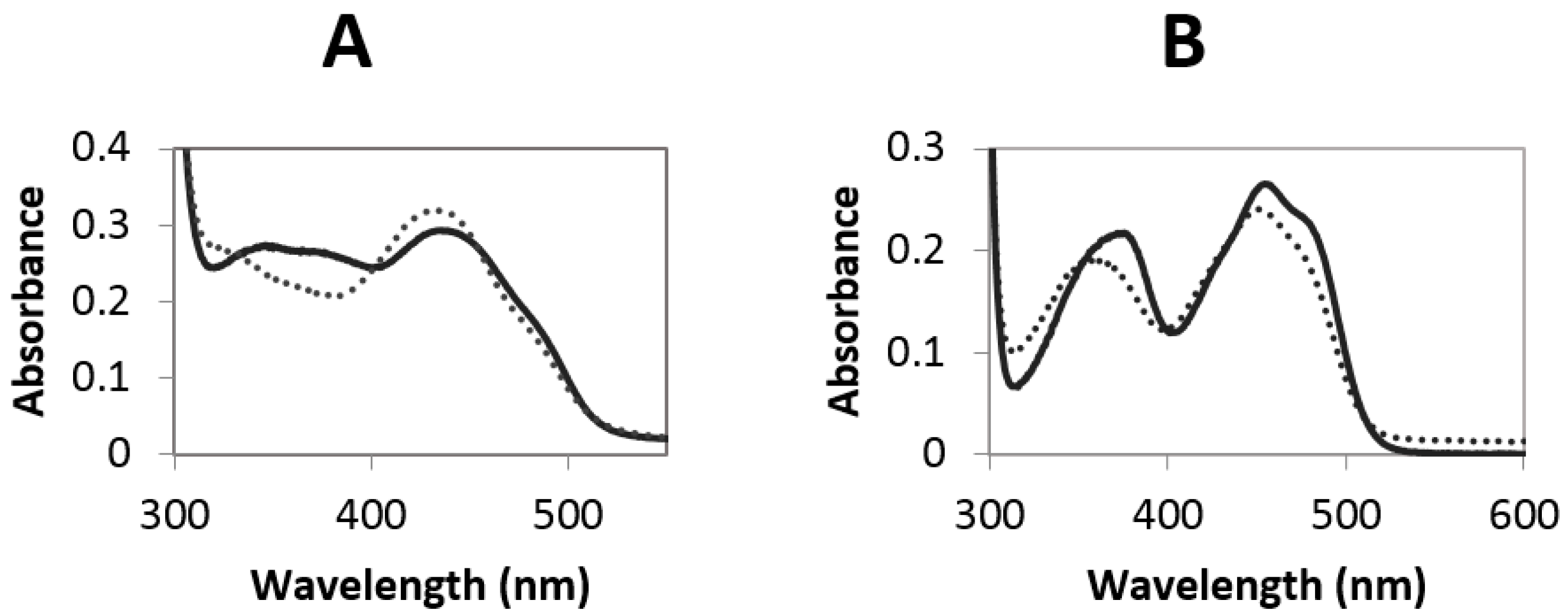
2.3. Redox Potential Determination and Stopped Flow Experiments
2.4. Substrate Screening
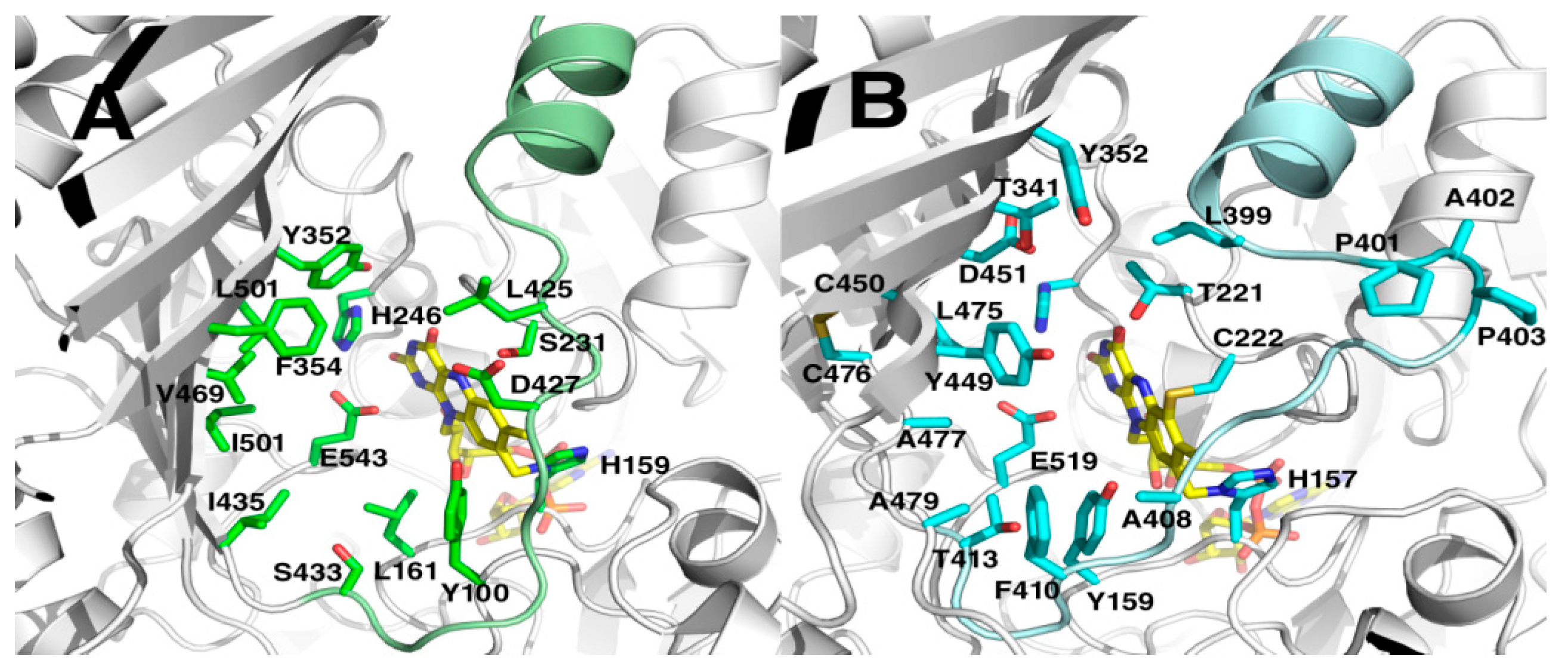
2.5. Crystal Structures
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis and Expression
4.2. Purification
4.3. Crystallization, Data Collection and Structure Determination of MtVAO615
4.4. Crystallization, Data Collection and Structure Determination of MtVAO713
4.5. Refinement
4.6. Carbohydrates and Primary Alcohol Substrate Screening
4.7. Steroids and Secondary Alcohol Substrate Screening
4.8. Stopped Flow Analysis
4.9. Redox Potential Determination
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dijkman, W.P.; de Gonzalo, G.; Mattevi, A.; Fraaije, M.W. Flavoprotein oxidases: Classification and applications. Appl. Microbiol. Biotechnol. 2013, 97, 5177–5188. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, M.W.; Van Berkel, W.J.; Benen, J.A.; Visser, J.; Mattevi, A. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem. Sci. 1998, 23, 206–207. [Google Scholar] [CrossRef]
- Xu, F.; Golightly, E.J.; Fuglsang, C.C.; Schneider, P.; Duke, K.R.; Lam, L.; Christensen, S.; Brown, K.M.; Jørgensen, C.T.; Brown, S.H. A novel carbohydrate: Acceptor oxidoreductase from Microdochium nivale. Eur. J. Biochem. 2001, 268, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Lai, W.-L.; Lee, M.-H.; Chen, C.-J.; Vasella, A.; Tsai, Y.-C.; Liaw, S.-H. Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: A novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD. J. Biol. Chem. 2005, 280, 38831–38838. [Google Scholar] [CrossRef] [PubMed]
- Heuts, D.P.H.M.; Scrutton, N.S.; McIntire, W.S.; Fraaije, M.W. What’s in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 2009, 276, 3405–3427. [Google Scholar] [CrossRef] [PubMed]
- Ewing, T.A.; Fraaije, M.W.; Mattevi, A.; van Berkel, W.J.H. The VAO/PCMH flavoprotein family. Arch Biochem. Biophys. 2017, 632, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.S.; Ho, J.Y.; Huang, C.C.; Lyu, S.Y.; Lee, C.Y.; Huang, Y.T.; Wu, C.J.; Chan, H.C.; Huang, C.J.; Hsu, N.S.; et al. A unique flavin mononucleotide-linked primary alcohol oxidase for glycopeptide A40926 maturation. J. Am. Chem. Soc. 2007, 129, 13384–13385. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, I.; Sultana, A.; Mäntsälä, P.; Niemi, J.; Schneider, G. Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc. Natl. Acad. Sci. USA 2007, 104, 6170–6175. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.C.; Li, S.; Gunatilleke, S.S.; Anzai, Y.; Burr, D.A.; Podust, L.M.; Sherman, D.H. Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat. Chem. 2011, 3, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.R.; Rozeboom, H.J.; Dobruchowska, J.M.; Van Leeuwen, S.S.; Vugts, A.S.C.; Koetsier, M.J.; Visser, J.; Fraaije, M.W.; Hart, G. Discovery of a xylooligosaccharide oxidase from Myceliophthora thermophila C1. J. Biol. Chem. 2016, 291, 23709–23718. [Google Scholar] [CrossRef] [PubMed]
- Heuts, D.P.H.M.; Janssen, D.B.; Fraaije, M.W. Changing the substrate specificity of a chitooligosaccharide oxidase from Fusarium graminearum by model-inspired site-directed mutagenesis. FEBS Lett. 2007, 581, 4905–4909. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yang, T.; Inukai, T.; Yamasaki, M.; Tsai, Y. Purification and characterization of a novel glucooligosaccharide oxidase from Acremonium stricture T1. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 1991, 18, 41–47. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Veeger, C.; van Berkel, W.J. Substrate specificity of flavin-dependent vanillyl-alcohol oxidase from Penicillium simplicissimum. Evidence for the production of 4-hydroxycinnamyl alcohols from 4-allylphenols. Eur. J. Biochem. 1995, 234, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Winter, R.T.; Fraaije, M.W. Applications of flavoprotein oxidases in organic synthesis—Novel reactivities that go beyond amine and alcohol oxidations. Curr. Org. Chem. 2012, 16, 2542–2550. [Google Scholar] [CrossRef]
- Winkler, A.; Hartner, F.; Kutchan, T.M.; Glieder, A.; Macheroux, P. Biochemical evidence that berberine bridge enzyme belongs to a novel family of flavoproteins containing a bi-covalently attached FAD cofactor. J. Biol. Chem. 2006, 281, 21276–21285. [Google Scholar] [CrossRef] [PubMed]
- Massey, V. A simple method for determination of redox potentials. In Flavins and Flavoproteins 1990; Curti, B., Ronchi, S., Zanetti, G., Eds.; John Wiley, Sons, Ltd.: Hoboken, NJ, USA, 1991; xxiii + 945pp. [Google Scholar]
- Mattevi, A. To be or not to be an oxidase: Challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 2006, 31, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Reeves, C.D.; Hu, Z.; Reid, R.; Kealey, J.T. Genes for the biosynthesis of the fungal polyketides hypothemycin from Hypomyces subiculosus and radicicol from Pochonia chlamydosporia. Appl. Environ. Microbiol. 2008, 74, 5121–5129. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, M.; Saito, H.; Niwa, R.; Niimi, T.; Toyoda, K.; Ueno, C.; Kanamori, Y.; Shimura, S.; Kiuchi, M. Fungal ecdysteroid-22-oxidase, a new tool for manipulating ecdysteroid signaling and insect development. J. Biol. Chem. 2012, 287, 16488–16498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.-H.; Lee, S.-B.; Kim, H.-S.; Jeong, K.-J.; Park, J.-Y.; Park, K.-M.; Lee, J.-W. Whole cell bioconversion of ricinoleic acid to 12-ketooleic acid by recombinant Corynebacterium glutamicum-based biocatalyst. J. Microbiol. Biotechnol. 2015, 25, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Leferink, N.G.H.; Heuts, D.P.H.M.; Fraaije, M.W.; van Berkel, W.J.H. The growing VAO flavoprotein family. Arch. Biochem. Biophys. 2008, 474, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Miyanaga, A.; Michaudel, Q.; Stull, F.; Louie, G.; Noel, J.P.; Baran, P.S.; Palfey, B.; Moore, B.S. Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement. Nature 2013, 503, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Koetter, J.W.A.; Schulz, G.E. Crystal structure of 6-hydroxy-d-nicotine oxidase from Arthrobacter nicotinovorans. J. Mol. Biol. 2005, 352, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Noinaj, N.; Bosserman, M.A.; Schickli, M.A.; Piszczek, G.; Kharel, M.K.; Pahari, P.; Buchanan, S.K.; Rohr, J. The crystal structure and mechanism of an unusual oxidoreductase, GilR, involved in gilvocarcin V biosynthesis. J. Biol. Chem. 2011, 286, 23533–23543. [Google Scholar] [CrossRef] [PubMed]
- Shoyama, Y.; Tamada, T.; Kurihara, K.; Takeuchi, A.; Taura, F.; Arai, S.; Blaber, M.; Shoyama, Y.; Morimoto, S.; Kuroki, R. Structure and function of ∆1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa. J. Mol. Biol. 2012, 423, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Zafred, D.; Steiner, B.; Teufelberger, A.R.; Hromic, A.; Karplus, P.A.; Schofield, C.J.; Wallner, S.; Macheroux, P. Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction. FEBS J. 2015, 282, 3060–3074. [Google Scholar] [CrossRef] [PubMed]
- Battye, T.G.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G.W. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Canutescu, A.A.; Shelenkov, A.A.; Dunbrack, R.L. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003, 12, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.-H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Métoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.R.; Lee, M.; Fraaije, M.W. Expanding the substrate scope of chitooligosaccharide oxidase from Fusarium graminearum by structure-inspired mutagenesis. Biotechnol. Bioeng. 2015, 112, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Minnaert, K. Measurement of the equilibrium constant of the reaction between cytochrome c and cytochrome a. Biochim. Biophys. Acta 1965, 110, 42–56. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
| MtVAO615 | MtVAO615 | MtVAO713 | |
|---|---|---|---|
| Diffraction data | pH 7.5 | pH 5.0 | |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 |
| Resolution range (Å) | 51.4–2.0 | 58.0–2.2 | 54.3–2.2 |
| Spacegroup | P212121 | P22121 | P21 |
| Cell dimensions (Å) a, b, c, β | 59.7, 100.9, 111.8 | 62.4, 116.0, 198.7 | 83.2, 108.5, 136.0, 90.0 |
| Number of unique reflections | 45,720 (2754) | 71,982 (4265) | 120,924 (5895) |
| Completeness (%) | 98.1 (81.1) | 99.5 (92.8) | 98.8 (97.3) |
| Overall I/σ (I) | 4.7 (1.0) | 12.9 (3.7) | 3.9 (1.6) |
| Rmerge (%) | 17.2 (98.6) | 14.7 (51.4) | 16.1 (48.3) |
| Rpim (%) | 11.4 (70.7) | 5.8 (22.5) | 14.7 (47.9) |
| R/Rfree (%) | 23.6/28.0 | 19.5/23.5 | 19.8/23.9 |
| R.m.s. deviations from ideal values | |||
| Bond lengths (Å) | 0.010 | 0.009 | 0.013 |
| Bond angles (°) | 1.444 | 1.333 | 1.575 |
| Protein residues | 27–475 | 27–475 (both molecules) | 30–598 (4 molecules) |
| FAD molecule | 1 | 2 × 1 | 4 × 1 |
| Water molecules | 366 | 850 | 1547 |
| Glycerol molecules | - | 8 | |
| NAG | 9 | 4 (A), 7 (B) | 4 × 3 |
| β-mannose | 1 | - | - |
| PDB accession ID | 6F72 | 6F73 | 6F74 |
| Position Relative to MtVAO615 | 159 | 237 | 451 | 475 | 477 | 519 | ||
|---|---|---|---|---|---|---|---|---|
| 1DII | F | V | I | H | V | Y (?) | ||
| 1VAO | Y | S | V | T | H | I | Y (?) | |
| 5FXD | G | V | I | H | V | Y (?) | ||
| MtVAO713 | L | H | L | V | I | E | ||
| MtVAO615 | Y | H | D | L | A | E | ||
| 3POP | G | Y | N | K | S | Y | ||
| 2IPI | F | Y | Y | K | W | Y | ||
| 5AWV | F | Y | I | K | N | Y | ||
| 2Y08 | Y | Y | I | K | V | Y | ||
| 3D2D | Y | F | N | M | E | H | ||
| 3VTE | A | Y | Y | E | W | Y | ||
| 4UD8 | Y | Y | N | K | Q | Y | ||
| 3TSH | Y | F | D | N | Q | Y | ||
| 4DNS | Y | F | D | N | Q | Y | ||
| 3RJ8 | Y | F | D | L | Q | Y | ||
| 5L6F | Y | F | D | L | E | Y | ||
| 2AXR | Y | Y | D | L | Q | Y | ||
| ChitO | Y | Y | D | L | Q | Y | ||
| 2BVF | P | V | E | E | L | N | ||
| 3W8W | M | F | L | V | N | F | ||
| 1I19 | W | H | R | W | N | K | E | |
| 2VFR | F | H | R | A | H | K | ||
| 4KW5 | Y | H | K | N | C | K | ||
| 4BBY | S | H | T | I | P | H (?) | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrari, A.R.; Rozeboom, H.J.; Vugts, A.S.C.; Koetsier, M.J.; Floor, R.; Fraaije, M.W. Characterization of Two VAO-Type Flavoprotein Oxidases from Myceliophthora thermophila. Molecules 2018, 23, 111. https://doi.org/10.3390/molecules23010111
Ferrari AR, Rozeboom HJ, Vugts ASC, Koetsier MJ, Floor R, Fraaije MW. Characterization of Two VAO-Type Flavoprotein Oxidases from Myceliophthora thermophila. Molecules. 2018; 23(1):111. https://doi.org/10.3390/molecules23010111
Chicago/Turabian StyleFerrari, Alessandro R., Henriëtte J. Rozeboom, Aniek S. C. Vugts, Martijn J. Koetsier, Robert Floor, and Marco W. Fraaije. 2018. "Characterization of Two VAO-Type Flavoprotein Oxidases from Myceliophthora thermophila" Molecules 23, no. 1: 111. https://doi.org/10.3390/molecules23010111





