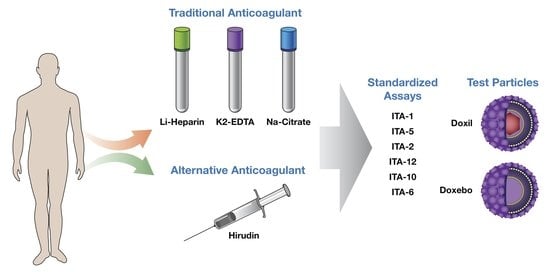
Anticoagulants Influence the Performance of In Vitro Assays Intended for Characterization of Nanotechnology-Based Formulations
Abstract
:1. Introduction
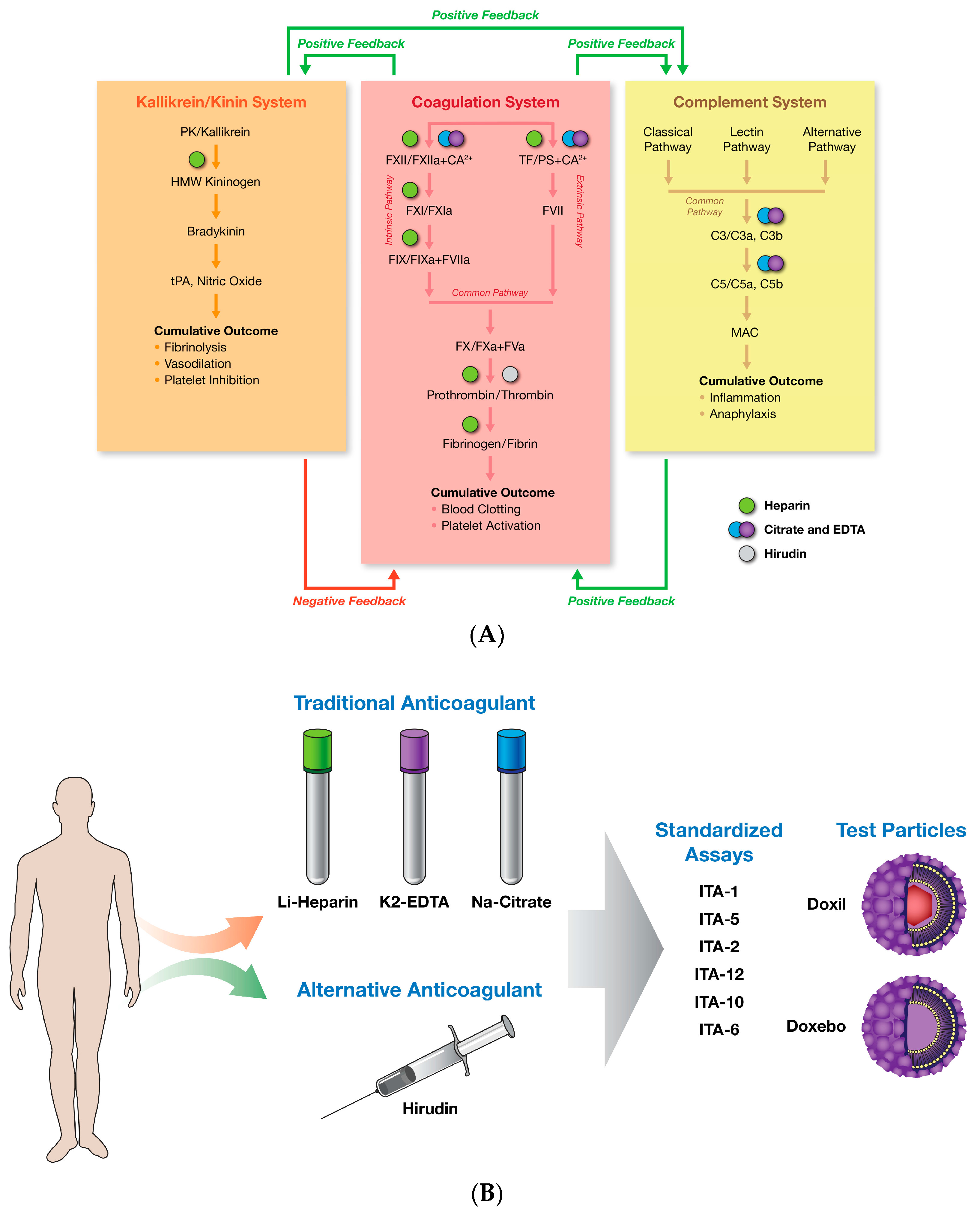
2. Results and Discussion
2.1. Physicochemical Characterization and Endotoxin
2.2. Hemolysis
2.3. Platelet Aggregation
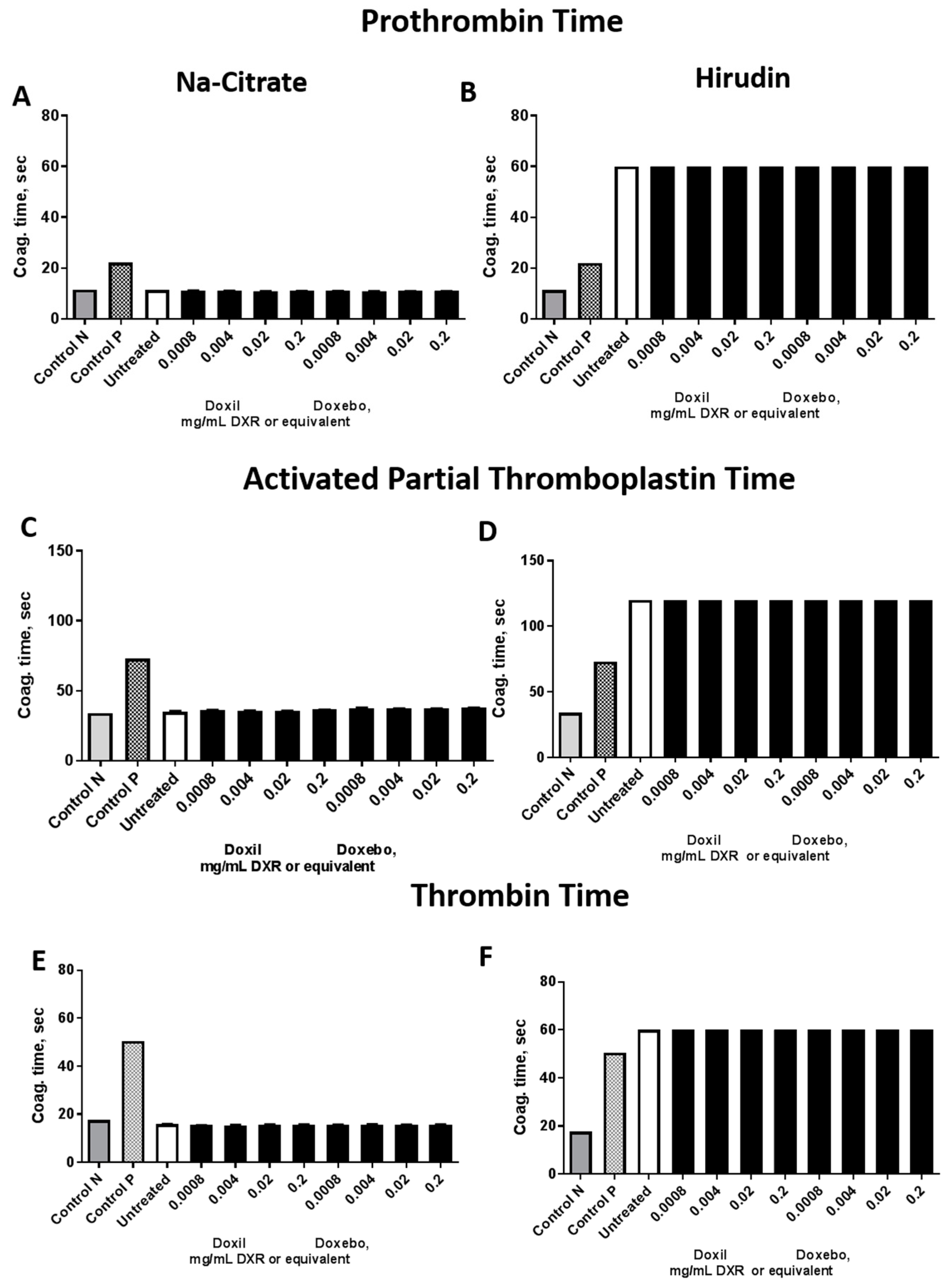
2.4. Plasma Coagulation Time
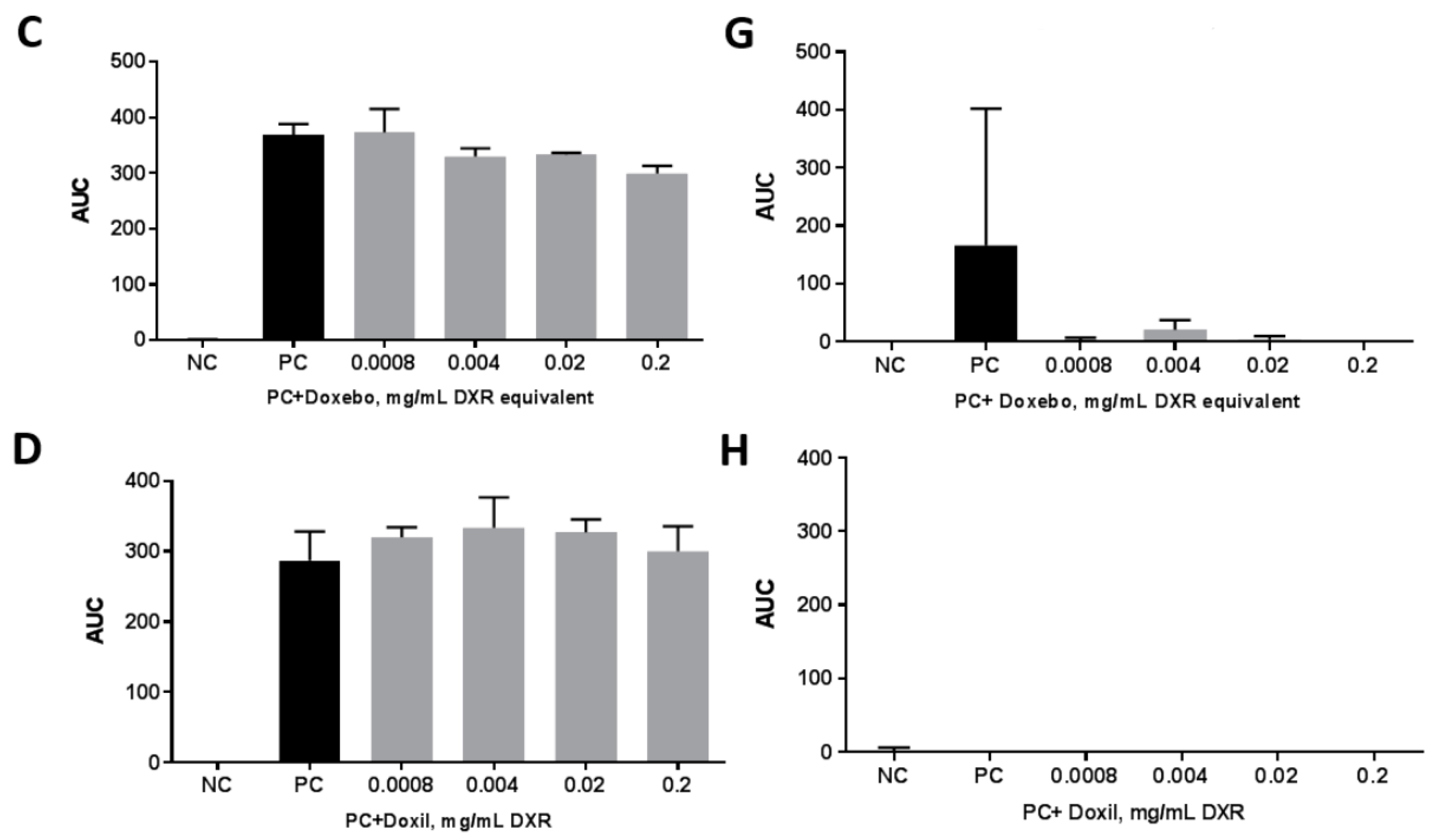
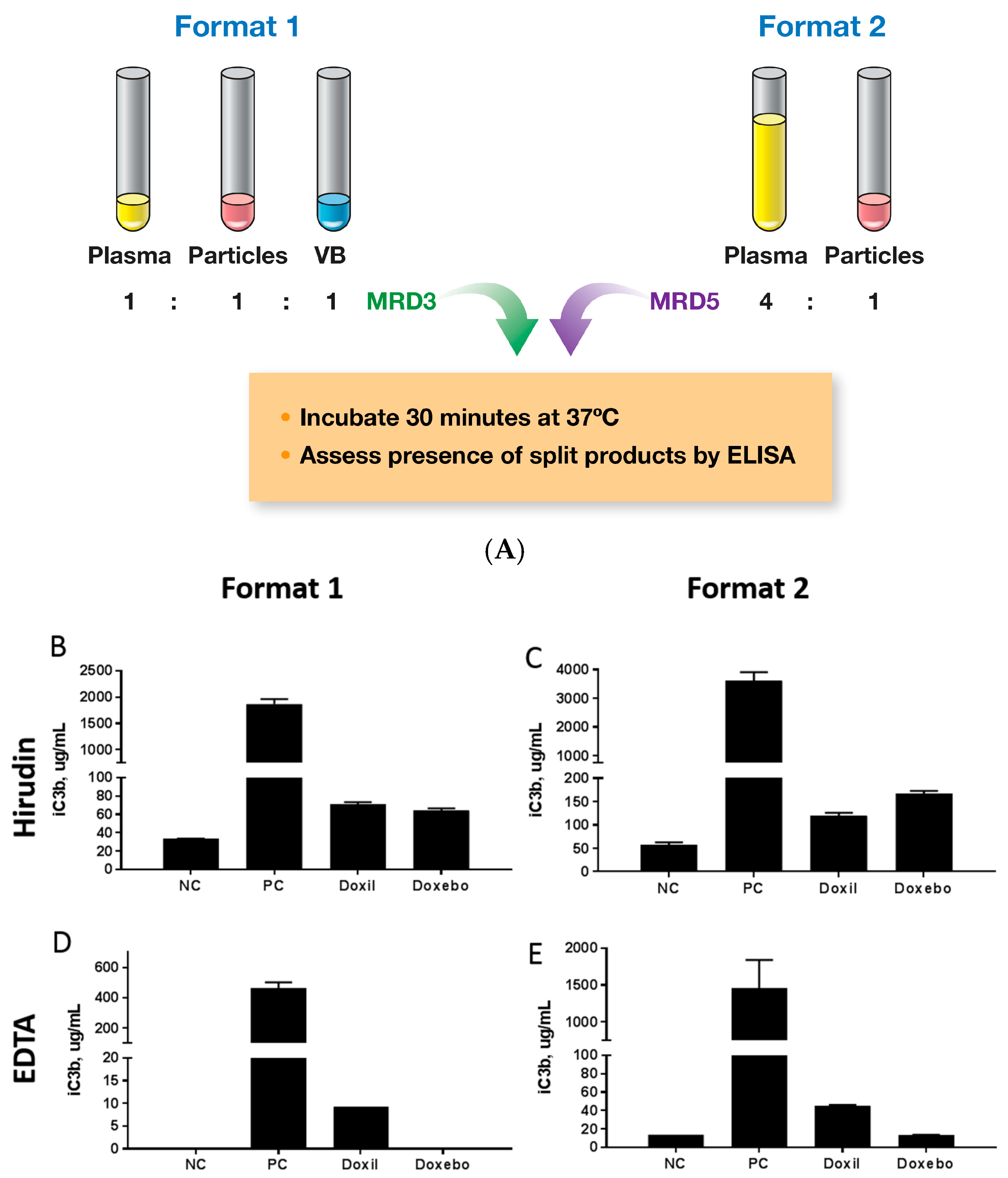
2.5. Complement Activation
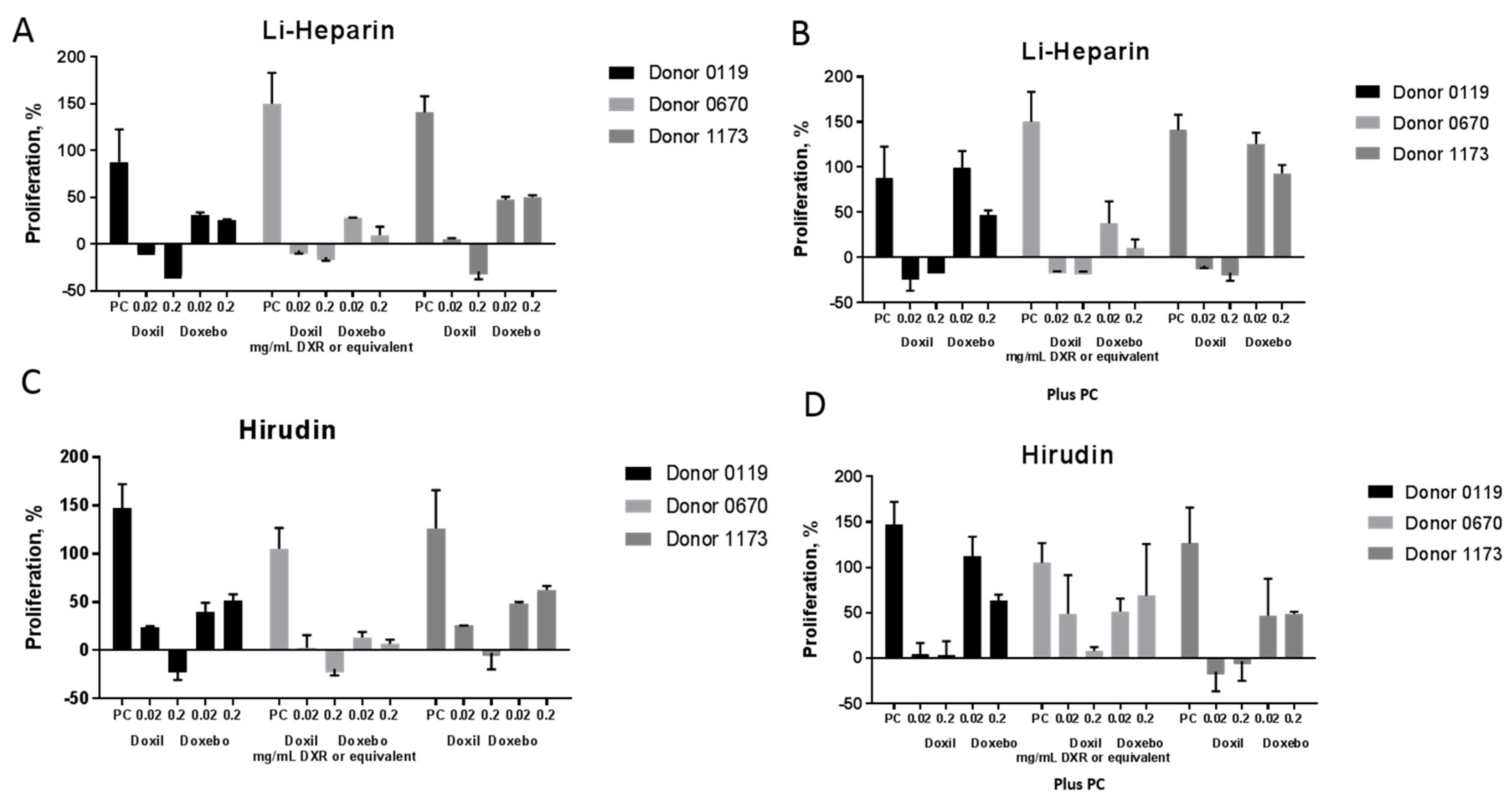
2.6. Leukocyte Proliferation
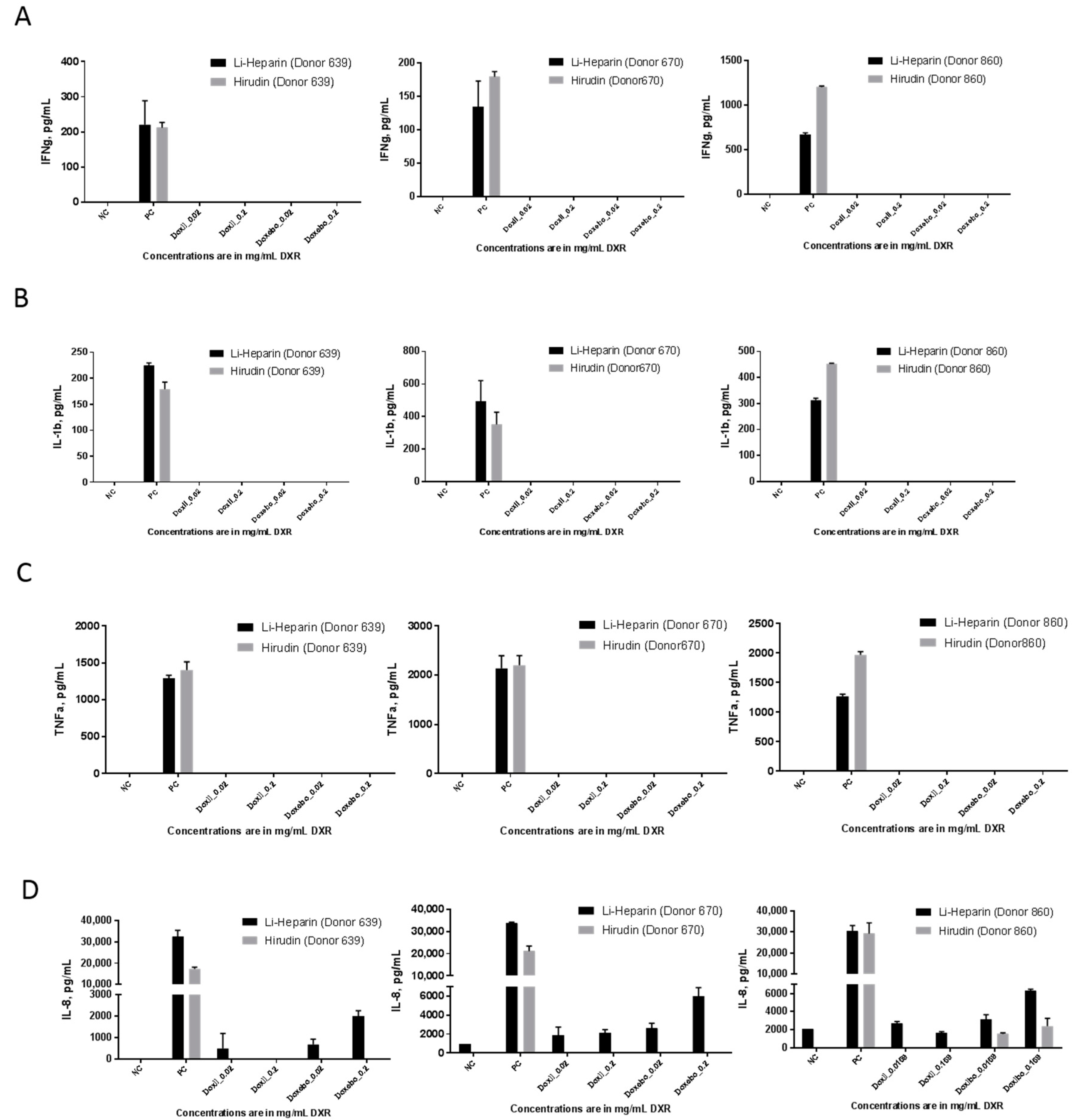
2.7. Cytokine Response
3. Materials and Methods
3.1. Reagents
3.2. DLS and Zeta Potential
3.3. Research Donor Blood
3.4. Endotoxin
3.5. Hemolysis
3.6. Platelet Aggregation
3.7. Complement Activation
3.8. Plasma Coagulation Time
3.9. Leukocyte Proliferation
3.10. Cytokine Response in PBMC Cultures
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Balveren, J.A.; Huijskens, M.J.; Gemen, E.F.; Pequeriaux, N.C.; Kusters, R. Effects of time and temperature on 48 routine chemistry, haematology and coagulation analytes in whole blood samples. Ann. Clin. Biochem. 2017, 54, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Wallen, N.H.; Ladjevardi, M.; Albert, J.; Broijersen, A. Influence of different anticoagulants on platelet aggregation in whole blood: A comparison between citrate, low molecular mass heparin and hirudin. Thromb. Res. 1997, 87, 151–157. [Google Scholar] [CrossRef]
- Kenne, E.; Renne, T. Factor XII: A drug target for safe interference with thrombosis and inflammation. Drug Discov. Today 2014, 19, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Bexborn, F.; Engberg, A.E.; Sandholm, K.; Mollnes, T.E.; Hong, J.; Ekdahl, K.N. Hirudin versus heparin for use in whole blood in vitro biocompatibility models. J. Biomed. Mater. Res. A 2009, 89, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Carteaux, J.P.; Gast, A.; Tschopp, T.B.; Roux, S. Activated clotting time as an appropriate test to compare heparin and direct thrombin inhibitors such as hirudin or Ro 46-6240 in experimental arterial thrombosis. Circulation 1995, 91, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.R.; Liu, Y.; Lu, W.L.; Zhao, J.H.; Wang, X.Q.; Zhang, H.; Wang, J.C.; Zhang, X.; Zhang, Q. A recombinant peptide, hirudin, potentiates the inhibitory effects of stealthy liposomal vinblastine on the growth and metastasis of melanoma. Biol. Pharm. Bull. 2008, 31, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Kapur, N.K.; Shenoy, C.; Yunis, A.A.; Mohammad, N.N.; Wilson, S.; Paruchuri, V.; Mackey, E.E.; Qiao, X.; Shah, A.; Esposito, M.L.; et al. Distinct effects of unfractionated heparin versus bivalirudin on circulating angiogenic peptides. PLoS ONE 2012, 7, e34344. [Google Scholar] [CrossRef] [PubMed]
- Kopp, R.; Bernsberg, R.; Kashefi, A.; Mottaghy, K.; Rossaint, R.; Kuhlen, R. Effect of hirudin versus heparin on hemocompatibility of blood contacting biomaterials: An in vitro study. Int. J. Artif. Organs 2005, 28, 1272–1277. [Google Scholar] [PubMed]
- Krishnan, R.; Tulinsky, A.; Vlasuk, G.P.; Pearson, D.; Vallar, P.; Bergum, P.; Brunck, T.K.; Ripka, W.C. Synthesis, structure, and structure-activity relationships of divalent thrombin inhibitors containing an alpha-keto-amide transition-state mimetic. Protein Sci. 1996, 5, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Lisman, T.; Weeterings, C.; de Groot, P.G. Platelet aggregation: Involvement of thrombin and fibrin(ogen). Front. Biosci. 2005, 10, 2504–2517. [Google Scholar] [CrossRef] [PubMed]
- Menssen, H.D.; Melber, K.; Brandt, N.; Thiel, E. The use of hirudin as universal anticoagulant in haematology, clinical chemistry and blood grouping. Clin. Chem. Lab. Med. 2001, 39, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, W.; Wang, L.; Chintala, M.; Plump, A.S.; Ogletree, M.L.; Chen, Z. Differential profiles of thrombin inhibitors (heparin, hirudin, bivalirudin, and dabigatran) in the thrombin generation assay and thromboelastography in vitro. Blood Coagul. Fibrinolysis 2013, 24, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Barkley, T.W., Jr. Medicinal leech therapy: New life for an ancient treatment. Nursing 2015, 45, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Porshinsky, B.S.; Saha, S.; Grossman, M.D.; Ii, P.R.B.; Stawicki, S.P. Clinical uses of the medicinal leech: A practical review. J. Postgrad. Med. 2011, 57, 65–71. [Google Scholar] [PubMed]
- Singh, A.P. Medicinal leech therapy (hirudotherapy): A brief overview. Complement. Ther. Clin. Pract. 2010, 16, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaia, M.A.; McNeil, S.E. Immunological properties of engineered nanomaterials. Nat. Nanotechnol. 2007, 2, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Barenholz, Y. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Staropoli, N.; Ciliberto, D.; Botta, C.; Fiorillo, L.; Grimaldi, A.; Lama, S.; Caraglia, M.; Salvino, A.; Tassone, P.; Tagliaferri, P. Pegylated liposomal doxorubicin in the management of ovarian cancer: A systematic review and metaanalysis of randomized trials. Cancer Biol. Ther. 2014, 15, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaia, M.A. Pre-clinical immunotoxicity studies of nanotechnology-formulated drugs: Challenges, considerations and strategy. J. Control. Release 2015, 220, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, G.E.; Evans, R.J.; Heath, M.F. The role of ADP in endotoxin-induced equine platelet activation. Eur. J. Pharmacol. 1996, 315, 203–212. [Google Scholar] [CrossRef]
- Szebeni, J.; Wassef, N.M.; Rudolph, A.S.; Alving, C.R. Complement activation by liposome-encapsulated hemoglobin in vitro: The role of endotoxin contamination. Artif. Cells Blood Substit. Immobil. Biotechnol. 1995, 23, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Engstad, C.S.; Gutteberg, T.J.; Osterud, B. Modulation of blood cell activation by four commonly used anticoagulants. Thromb. Haemost. 1997, 77, 690–696. [Google Scholar] [PubMed]
- Hauptmann, J.; Kaiser, B. Anticoagulant potential of synthetic and recombinant inhibitors of factor Xa and thrombin in vitro. Blood Coagul. Fibrinolysis 1993, 4, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, J. Complement activation-related pseudoallergy: A stress reaction in blood triggered by nanomedicines and biologicals. Mol. Immunol. 2014, 61, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, J.; Muggia, F.; Gabizon, A.; Barenholz, Y. Activation of complement by therapeutic liposomes and other lipid excipient-based therapeutic products: Prediction and prevention. Adv. Drug Deliv. Rev. 2011, 63, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, J.; Storm, G. Complement activation as a bioequivalence issue relevant to the development of generic liposomes and other nanoparticulate drugs. Biochem. Biophys. Res. Commun. 2015, 468, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Kozma, T.G.; Meszaros, T.; Weiszhar, Z.; Schneider, T.; Rosta, A.; Urbanics, R.; Rosivall, L.; Szebeni, J. Variable association of complement activation by rituximab and paclitaxel in cancer patients in vivo and in their screening serum in vitro with clinical manifestations of hypersensitivity: A pilot study. Eur. J. Nanomed. 2015, 7, 289–301. [Google Scholar] [CrossRef]
- Mollnes, T.E.; Brekke, O.L.; Fung, M.; Fure, H.; Christiansen, D.; Bergseth, G.; Videm, V.; Lappegard, K.T.; Kohl, J.; Lambris, J.D. Essential role of the C5a receptor in E coli-induced oxidative burst and phagocytosis revealed by a novel lepirudin-based human whole blood model of inflammation. Blood 2002, 100, 1869–1877. [Google Scholar] [PubMed]
- Menssen, H.D.; Brandt, N.; Leben, R.; Muller, F.; Thiel, E.; Melber, K. Measurement of hematological, clinical chemistry, and infection parameters from hirudinized blood collected in universal blood sampling tubes. Semin. Thromb. Hemost. 2001, 27, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Neun, B.W.; Dobrovolskaia, M.A. Detection and quantitative evaluation of endotoxin contamination in nanoparticle formulations by LAL-based assays. Methods Mol. Biol. 2011, 697, 121–130. [Google Scholar] [PubMed]
- Neun, B.W.; Dobrovolskaia, M.A. Considerations and Some Practical Solutions to Overcome Nanoparticle Interference with LAL Assays and to Avoid Endotoxin Contamination in Nanoformulations. Methods Mol. Biol. 2018, 1682, 23–33. [Google Scholar] [PubMed]
- Neun, B.W.; Ilinskaya, A.N.; Dobrovolskaia, M.A. Updated Method for In Vitro Analysis of Nanoparticle Hemolytic Properties. Methods Mol. Biol. 2018, 1682, 91–102. [Google Scholar] [PubMed]
- Potter, T.M.; Rodriguez, J.C.; Neun, B.W.; Ilinskaya, A.N.; Cedrone, E.; Dobrovolskaia, M.A. In Vitro Assessment of Nanoparticle Effects on Blood Coagulation. Methods Mol. Biol. 2018, 1682, 103–124. [Google Scholar] [PubMed]
- Neun, B.W.; Ilinskaya, A.N.; Dobrovolskaia, M.A. Analysis of Complement Activation by Nanoparticles. Methods Mol. Biol. 2018, 1682, 149–160. [Google Scholar] [PubMed]
- Potter, T.M.; Neun, B.W.; Dobrovolskaia, M.A. Methods for Analysis of Nanoparticle Immunosuppressive Properties In Vitro and In Vivo. Methods Mol. Biol. 2018, 1682, 161–172. [Google Scholar] [PubMed]
- Potter, T.M.; Neun, B.W.; Rodriguez, J.C.; Ilinskaya, A.N.; Dobrovolskaia, M.A. Analysis of Pro-inflammatory Cytokine and Type II Interferon Induction by Nanoparticles. Methods Mol. Biol. 2018, 1682, 173–187. [Google Scholar] [PubMed]
Sample Availability: Samples of the model nanoparticles Doxil and Doxebo are available from a commercial supplier. |


| Model Particles | Hydrodynamic Size (nm) | Zeta Potential (mV) | Drug (Conc. in mg/mL) | Endotoxin, EU/mL |
|---|---|---|---|---|
| Doxil | 87.8 | −4.0 | DXR (2.0) | <0.5 * |
| Doxebo | 83.1 | −4.0 | None | <0.5 ** |
| Anticoagulant | Main MOA | Targets outside of the Blood Coagulation | Assay Recommendation |
|---|---|---|---|
| EDTA | Chelation of divalent cations | yes | Complement activation * |
| Citrate | Chelation of divalent cations | yes | Platelet aggregation, Plasma Coagulation |
| Heparin | Inhibition of intrinsic, extrinsic and common coagulation pathways | yes | Hemolysis, Cytokines, Leukocyte proliferation * |
| Hirudin | Specific Thrombin inhibition | no | Complement activation, Hemolysis, Leukocyte Proliferation |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cedrone, E.; Neun, B.W.; Rodriguez, J.; Vermilya, A.; Clogston, J.D.; McNeil, S.E.; Barenholz, Y.; Szebeni, J.; Dobrovolskaia, M.A. Anticoagulants Influence the Performance of In Vitro Assays Intended for Characterization of Nanotechnology-Based Formulations. Molecules 2018, 23, 12. https://doi.org/10.3390/molecules23010012
Cedrone E, Neun BW, Rodriguez J, Vermilya A, Clogston JD, McNeil SE, Barenholz Y, Szebeni J, Dobrovolskaia MA. Anticoagulants Influence the Performance of In Vitro Assays Intended for Characterization of Nanotechnology-Based Formulations. Molecules. 2018; 23(1):12. https://doi.org/10.3390/molecules23010012
Chicago/Turabian StyleCedrone, Edward, Barry W. Neun, Jamie Rodriguez, Alison Vermilya, Jeffrey D. Clogston, Scott E. McNeil, Yechezkel Barenholz, Janos Szebeni, and Marina A. Dobrovolskaia. 2018. "Anticoagulants Influence the Performance of In Vitro Assays Intended for Characterization of Nanotechnology-Based Formulations" Molecules 23, no. 1: 12. https://doi.org/10.3390/molecules23010012