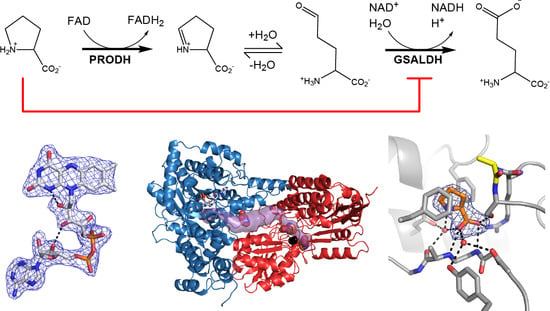
Structural Basis for the Substrate Inhibition of Proline Utilization A by Proline
Abstract
:1. Introduction
2. Results
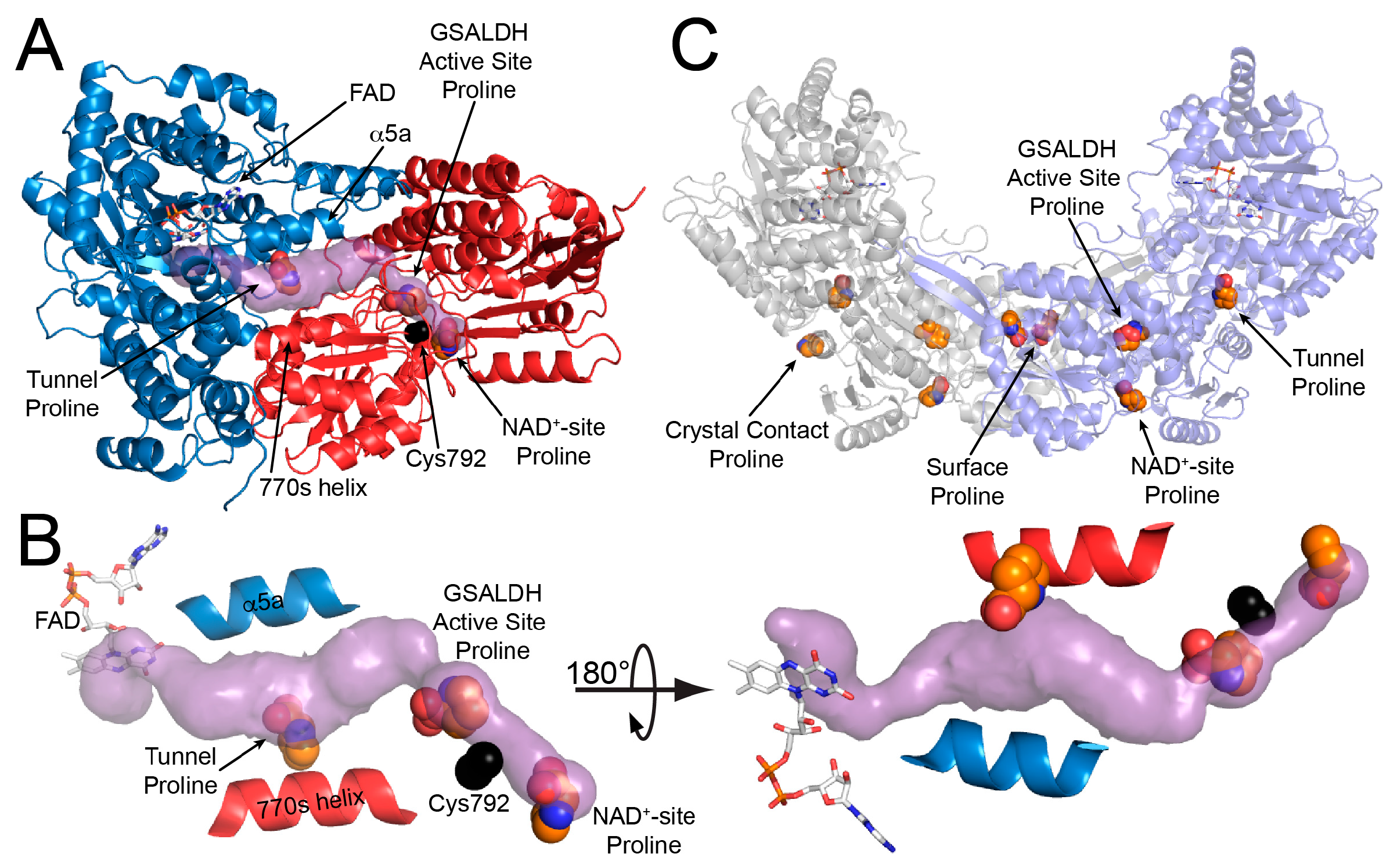
2.1. Overall Structure
2.2. FAD Conformation
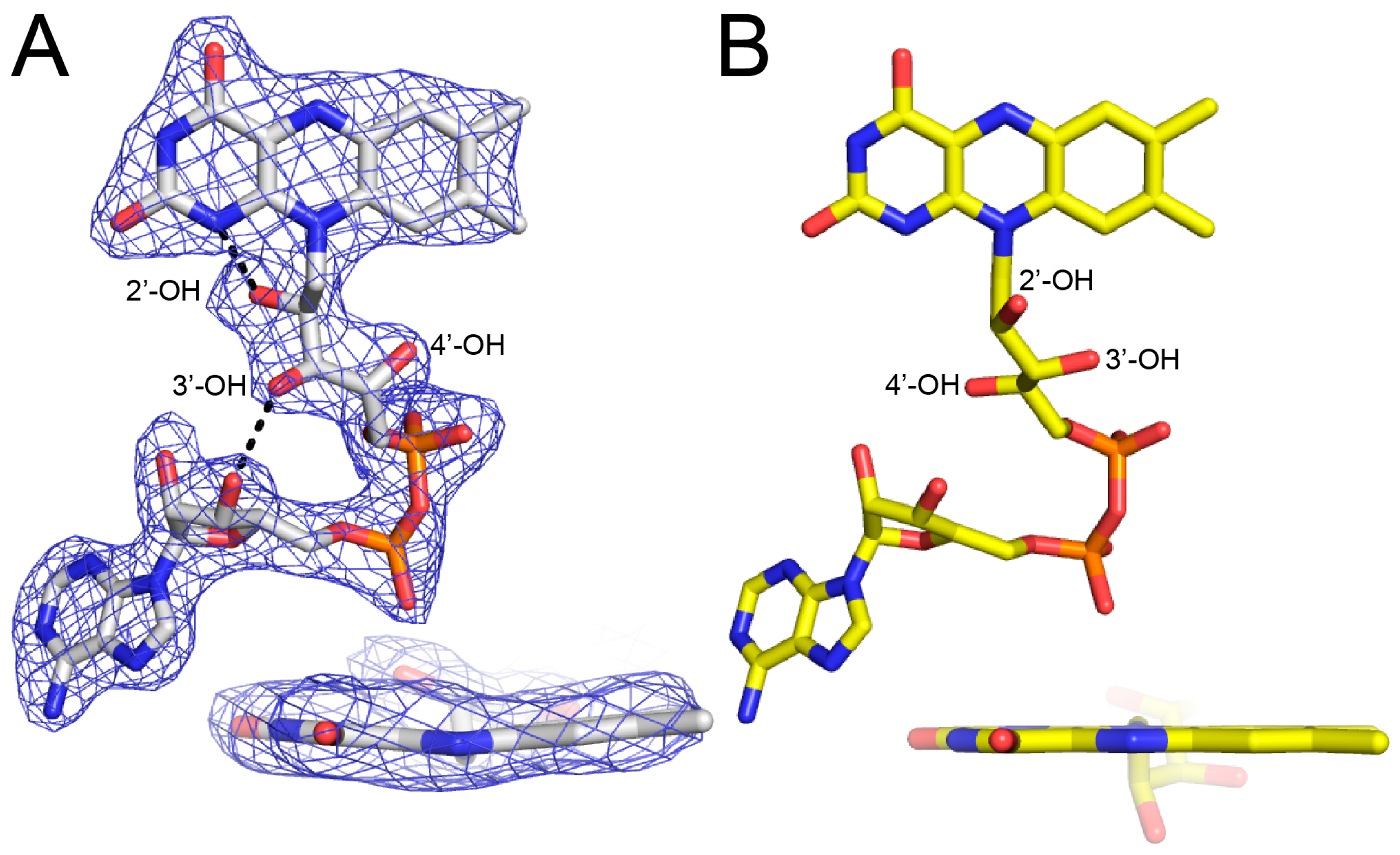
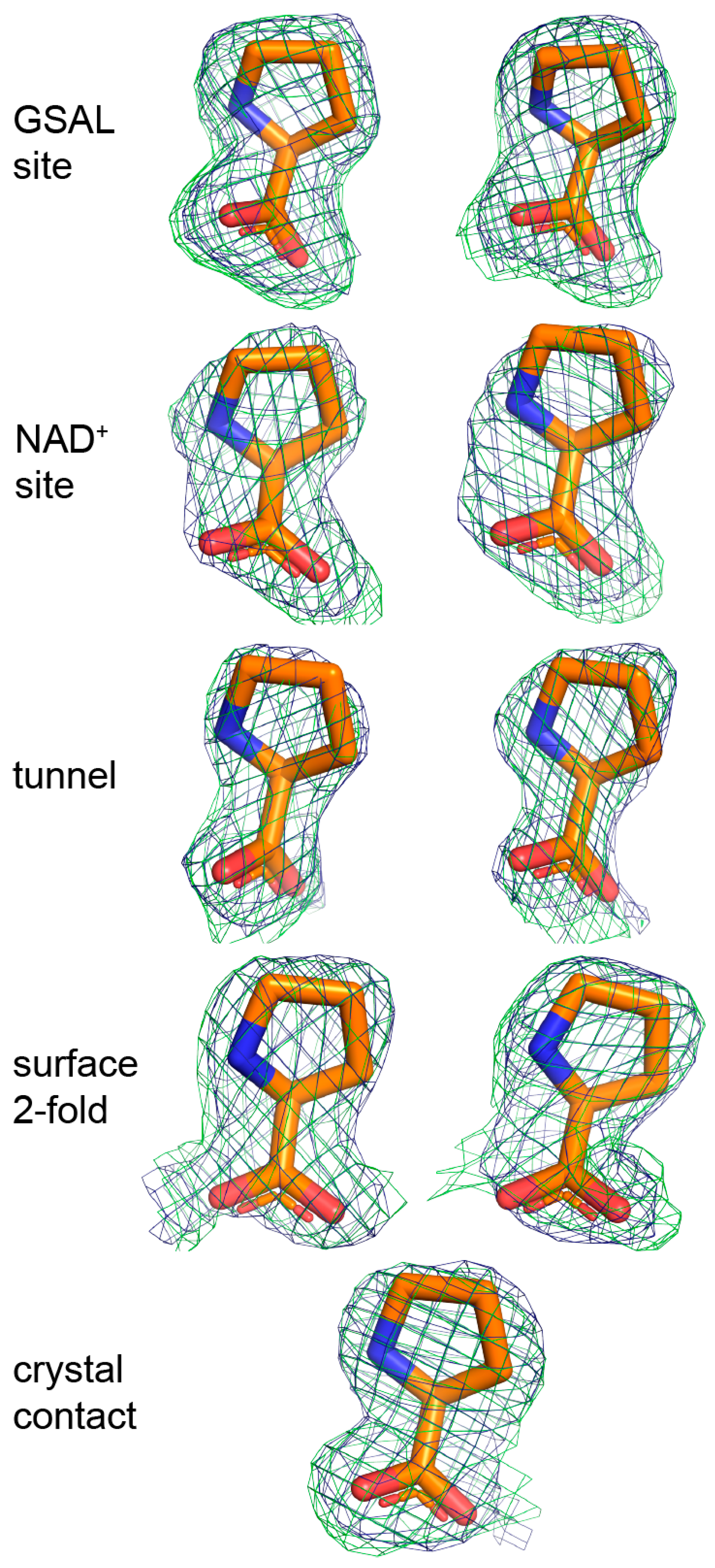
2.3. Proline Binding Sites
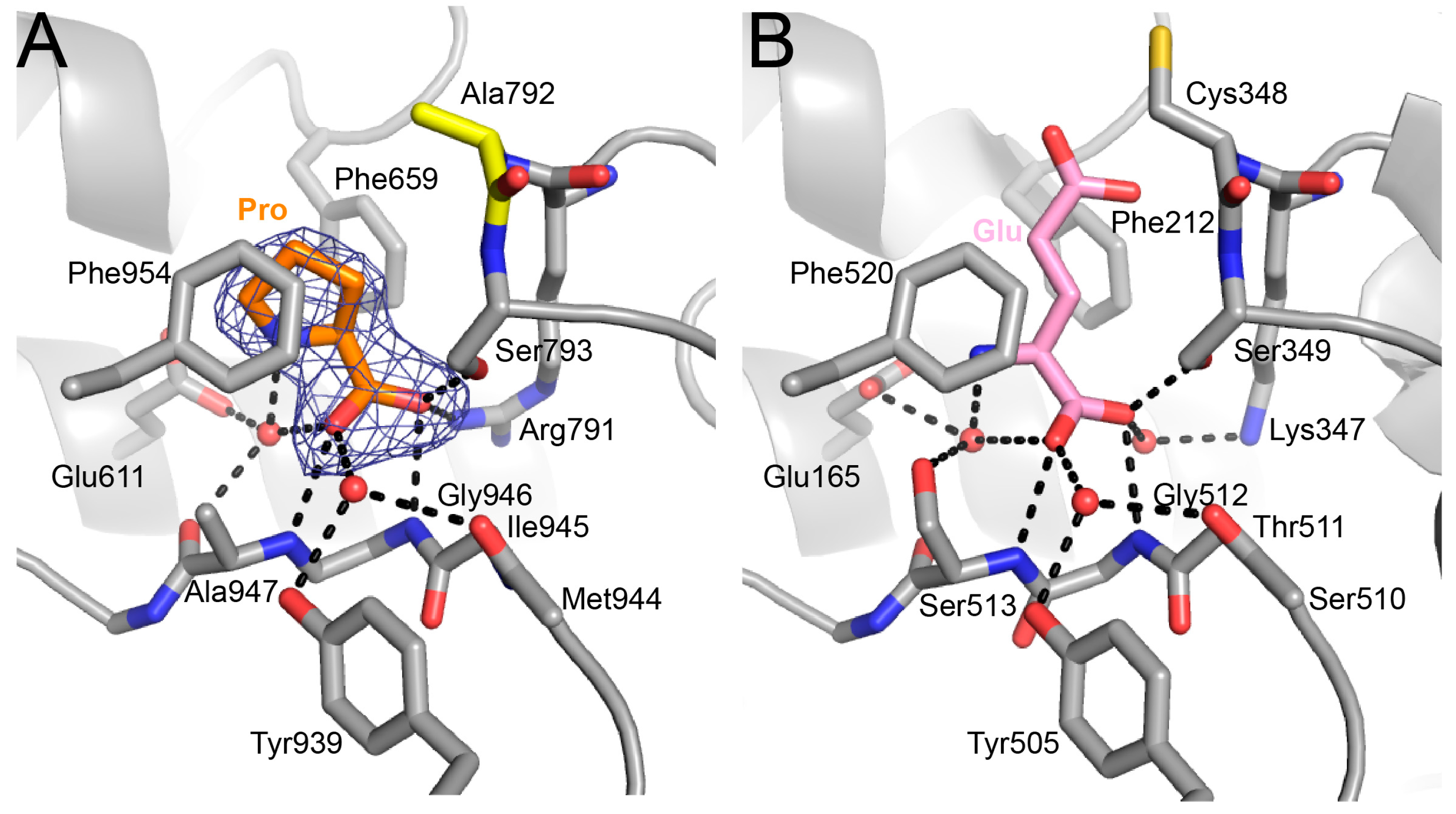
2.4. Proline Bound in the GSAL Site
2.5. Inhibition of Puta GSALDH Activity by Proline
3. Materials and Methods
3.1. X-ray Crystallography
3.2. Steady-State Kinetics Measurements
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, L.K.; Becker, D.F.; Tanner, J.J. Structure, function, and mechanism of proline utilization a (puta). Arch. Biochem. Biophys. 2017, 632, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Tanner, J.J. Structural biology of proline catabolic enzymes. Antioxid. Redox Signal. 2017. [Google Scholar] [CrossRef] [PubMed]
- Arentson, B.W.; Luo, M.; Pemberton, T.A.; Tanner, J.J.; Becker, D.F. Kinetic and structural characterization of tunnel-perturbing mutants in bradyrhizobium japonicum proline utilization a. Biochemistry 2014, 53, 5150–5161. [Google Scholar] [CrossRef] [PubMed]
- Moxley, M.A.; Sanyal, N.; Krishnan, N.; Tanner, J.J.; Becker, D.F. Evidence for hysteretic substrate channeling in the proline dehydrogenase and delta1-pyrroline-5-carboxylate dehydrogenase coupled reaction of proline utilization a (puta). J. Biol. Chem. 2014, 289, 3639–3651. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Gamage, T.T.; Arentson, B.W.; Schlasner, K.N.; Becker, D.F.; Tanner, J.J. Structures of proline utilization a (puta) reveal the fold and functions of the aldehyde dehydrogenase superfamily domain of unknown function. J. Biol. Chem. 2016, 291, 24065–24075. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Schuermann, J.P.; White, T.A.; Krishnan, N.; Sanyal, N.; Hura, G.L.; Tan, A.; Henzl, M.T.; Becker, D.F.; Tanner, J.J. Crystal structure of the bifunctional proline utilization a flavoenzyme from bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 2010, 107, 2878–2883. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Singh, H.; Pemberton, T.A.; Luo, M.; Dhatwalia, R.; Tanner, J.J. Biophysical investigation of type a putas reveals a conserved core oligomeric structure. FEBS J. 2017, 284, 3029–3049. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Arentson, B.W.; Becker, D.F.; Tanner, J.J. Structures of the puta peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc. Nat. Acad. Sci. USA 2014, 111, 3389–3394. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Zhu, W.; Johnson, W.H., Jr.; Whitman, C.P.; Becker, D.F.; Tanner, J.J. The structure of the proline utilization a proline dehydrogenase domain inactivated by n-propargylglycine provides insight into conformational changes induced by substrate binding and flavin reduction. Biochemistry 2010, 49, 560–569. [Google Scholar] [CrossRef] [PubMed]
- White, T.A.; Johnson, W.H., Jr.; Whitman, C.P.; Tanner, J.J. Structural basis for the inactivation of thermus thermophilus proline dehydrogenase by n-propargylglycine. Biochemistry 2008, 47, 5573–5580. [Google Scholar] [CrossRef] [PubMed]
- White, T.A.; Krishnan, N.; Becker, D.F.; Tanner, J.J. Structure and kinetics of monofunctional proline dehydrogenase from thermus thermophilus. J. Biol. Chem. 2007, 282, 14316–14327. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Arentson, B.W.; Srivastava, D.; Becker, D.F.; Tanner, J.J. Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release. Biochemistry 2012, 51, 10099–10108. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, T.A.; Still, B.R.; Christensen, E.M.; Singh, H.; Srivastava, D.; Tanner, J.J. Proline: Mother nature’s cryoprotectant applied to protein crystallography. Acta Crystallogr. D Struct. Biol. 2012, 68, 1010–1018. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Singh, R.K.; Moxley, M.A.; Henzl, M.T.; Becker, D.F.; Tanner, J.J. The three-dimensional structural basis of type ii hyperprolinemia. J. Mol. Biol. 2012, 420, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, E.; Ohshima, N.; Takahashi, H.; Kuroishi, C.; Yokoyama, S.; Tahirov, T.H. Crystal structure of thermus thermophilus delta1-pyrroline-5-carboxylate dehydrogenase. J. Mol. Biol. 2006, 362, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, T.A.; Tanner, J.J. Structural basis of substrate selectivity of delta(1)-pyrroline-5-carboxylate dehydrogenase (aldh4a1): Semialdehyde chain length. Arch. Biochem. Biophys. 2013, 538, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Tanner, J.J. Structural basis of substrate recognition by aldehyde dehydrogenase 7a1. Biochemistry 2015, 54, 5513–5522. [Google Scholar] [CrossRef] [PubMed]
- Riveros-Rosas, H.; Gonzalez-Segura, L.; Julian-Sanchez, A.; Diaz-Sanchez, A.G.; Munoz-Clares, R.A. Structural determinants of substrate specificity in aldehyde dehydrogenases. Chem. Biol. Interact. 2013, 202, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Nadaraia, S.; Gu, D.; Becker, D.F.; Tanner, J.J. Structure of the proline dehydrogenase domain of the multifunctional puta flavoprotein. Nat. Struct. Biol. 2003, 10, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; White, T.A.; Schuermann, J.P.; Baban, B.A.; Becker, D.F.; Tanner, J.J. Structures of the escherichia coli puta proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry 2004, 43, 12539–12548. [Google Scholar] [CrossRef] [PubMed]
- Ostrander, E.L.; Larson, J.D.; Schuermann, J.P.; Tanner, J.J. A conserved active site tyrosine residue of proline dehydrogenase helps enforce the preference for proline over hydroxyproline as the substrate. Biochemistry 2009, 48, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Schuermann, J.P.; White, T.A.; Srivastava, D.; Karr, D.B.; Tanner, J.J. Three crystal forms of the bifunctional enzyme proline utilization a (puta) from bradyrhizobium japonicum. Acta Crystallogr. F Struct. Biol. Commun. 2008, F64, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Struct. Biol. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Struct. Biol. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.Refine. Acta Crystallogr. D Struct. Biol. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of coot. Acta Crystallogr. D Struct. Biol. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., III; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. Molprobity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Struct. Biol. 2010, 66, 12–21. [Google Scholar]
- Williams, I.; Frank, L. Improved chemical synthesis and enzymatic assay of delta-1-pyrroline-5-carboxylic acid. Anal. Biochem. 1975, 64, 85–97. [Google Scholar] [CrossRef]
- Moxley, M.A.; Tanner, J.J.; Becker, D.F. Steady-state kinetic mechanism of the proline:Ubiquinone oxidoreductase activity of proline utilization a (puta) from escherichia coli. Arch. Biochem. Biophys. 2011, 516, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Forte-McRobbie, C.; Pietruszko, R. Human glutamic-gamma-semialdehyde dehydrogenase. Kinetic mechanism. Biochem. J. 1989, 261, 935–943. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples are not available from the authors. |


| Space Group | C2 |
|---|---|
| Unit cell parameters (Å, °) | a = 166.8 b = 194.2 c = 108.7 β = 121.4 |
| Wavelength | 1.000 |
| Resolution (Å) | 47.02–2.15 (2.19–2.15)a |
| Observations | 587,377 (21,732) |
| Unique reflections | 158,884 (7,418) |
| Rmerge(I) | 0.089 (0.787) |
| Rmeas(I) | 0.105 (0.975) |
| Rpim(I) | 0.054 (0.563) |
| Mean I/σ | 8.7 (1.7) |
| Mean CC1/2 | 0.996 (0.649) |
| Completeness (%) | 99.5 (94.1) |
| Multiplicity | 3.7 (2.9) |
| No. protein residues | 1949 |
| No. of atoms | |
| Protein | 14540 |
| FAD | 106 |
| Proline | 72 |
| Sulfate ions | 70 |
| Water | 719 |
| Rwork | 0.209 (0.320) |
| Rfree b | 0.239 (0.379) |
| RMSD bond lengths (Å) | 0.007 |
| RMSD bond angles (°) | 0.890 |
| Ramachandran plot c | |
| Favored (%) | 98.39 |
| Outliers (%) | 0.05 |
| Clashscore (PR) c | 2.69 (99%) |
| MolProbity score (PR) c | 1.31 (99%) |
| Average B-factor (Å2) | |
| Protein | 39.6 |
| FAD | 32.9 |
| Proline | 48.1 |
| Sulfate ions | 79.4 |
| Water | 37.3 |
| Coordinate error (Å) d | 0.28 |
| PDB ID | 6BSN |
| Site | B-Factor (Å2) | Occupancy | Hydrogen Bonds to the Protein |
|---|---|---|---|
| GSAL site | 35.8 | 1.00 | 4 |
| GSAL site | 35.3 | 1.00 | 4 |
| NAD+ site | 44.1 | 0.89 | 1 |
| NAD+ site | 55.8 | 0.95 | 1 |
| Middle of the tunnel | 54.6 | 0.89 | 0 |
| Middle of the tunnel | 61.5 | 0.90 | 0 |
| Surface, near the dimer 2-fold axis | 47.6 | 0.89 | 3 |
| Surface, near the dimer 2-fold axis | 49.8 | 0.92 | 3 |
| Crystal contact | 48.1 | 0.95 | 1 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korasick, D.A.; Pemberton, T.A.; Arentson, B.W.; Becker, D.F.; Tanner, J.J. Structural Basis for the Substrate Inhibition of Proline Utilization A by Proline. Molecules 2018, 23, 32. https://doi.org/10.3390/molecules23010032
Korasick DA, Pemberton TA, Arentson BW, Becker DF, Tanner JJ. Structural Basis for the Substrate Inhibition of Proline Utilization A by Proline. Molecules. 2018; 23(1):32. https://doi.org/10.3390/molecules23010032
Chicago/Turabian StyleKorasick, David A., Travis A. Pemberton, Benjamin W. Arentson, Donald F. Becker, and John J. Tanner. 2018. "Structural Basis for the Substrate Inhibition of Proline Utilization A by Proline" Molecules 23, no. 1: 32. https://doi.org/10.3390/molecules23010032