Exogenous 24-Epibrassinolide Interacts with Light to Regulate Anthocyanin and Proanthocyanidin Biosynthesis in Cabernet Sauvignon (Vitis vinifera L.)
Abstract
:1. Introduction
2. Results
2.1. Physiochemical Parameters
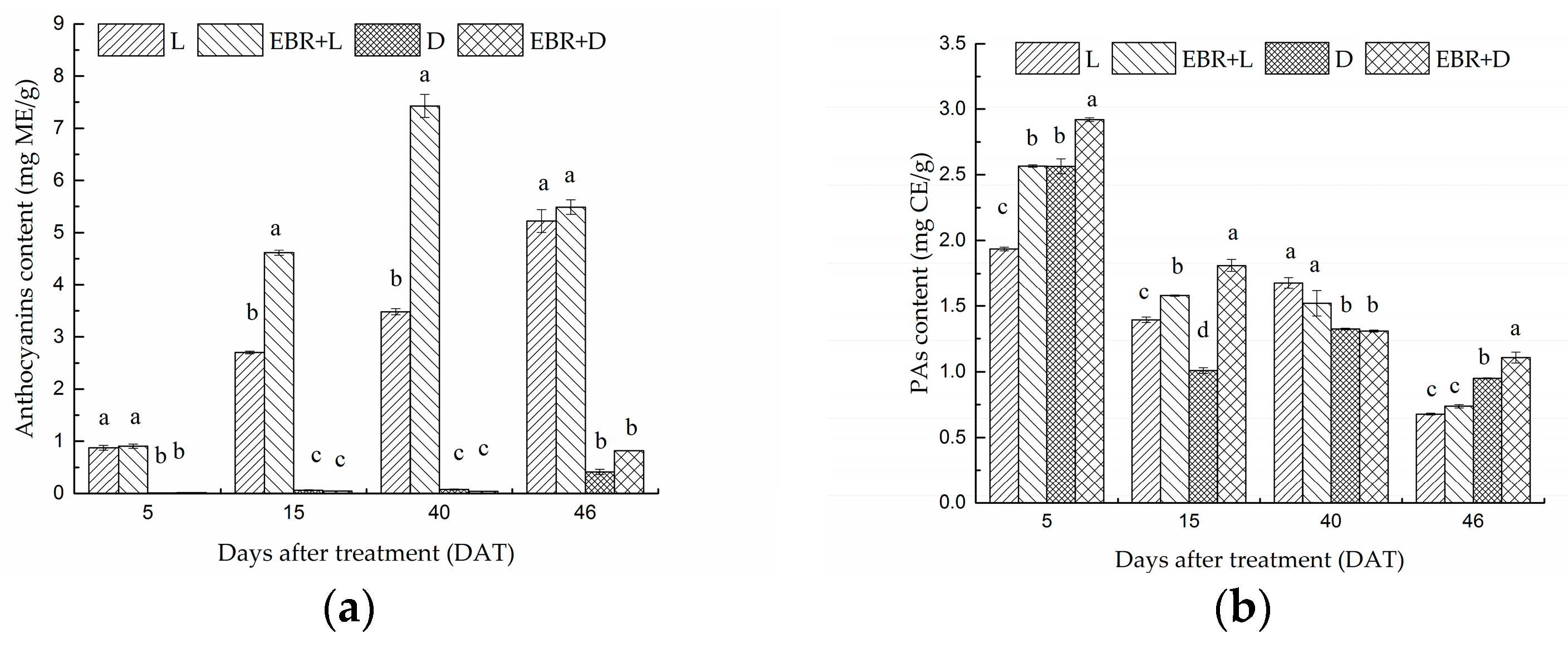
2.2. The Content of Anthocyanins and Proanthocyanidins in Grape Skins
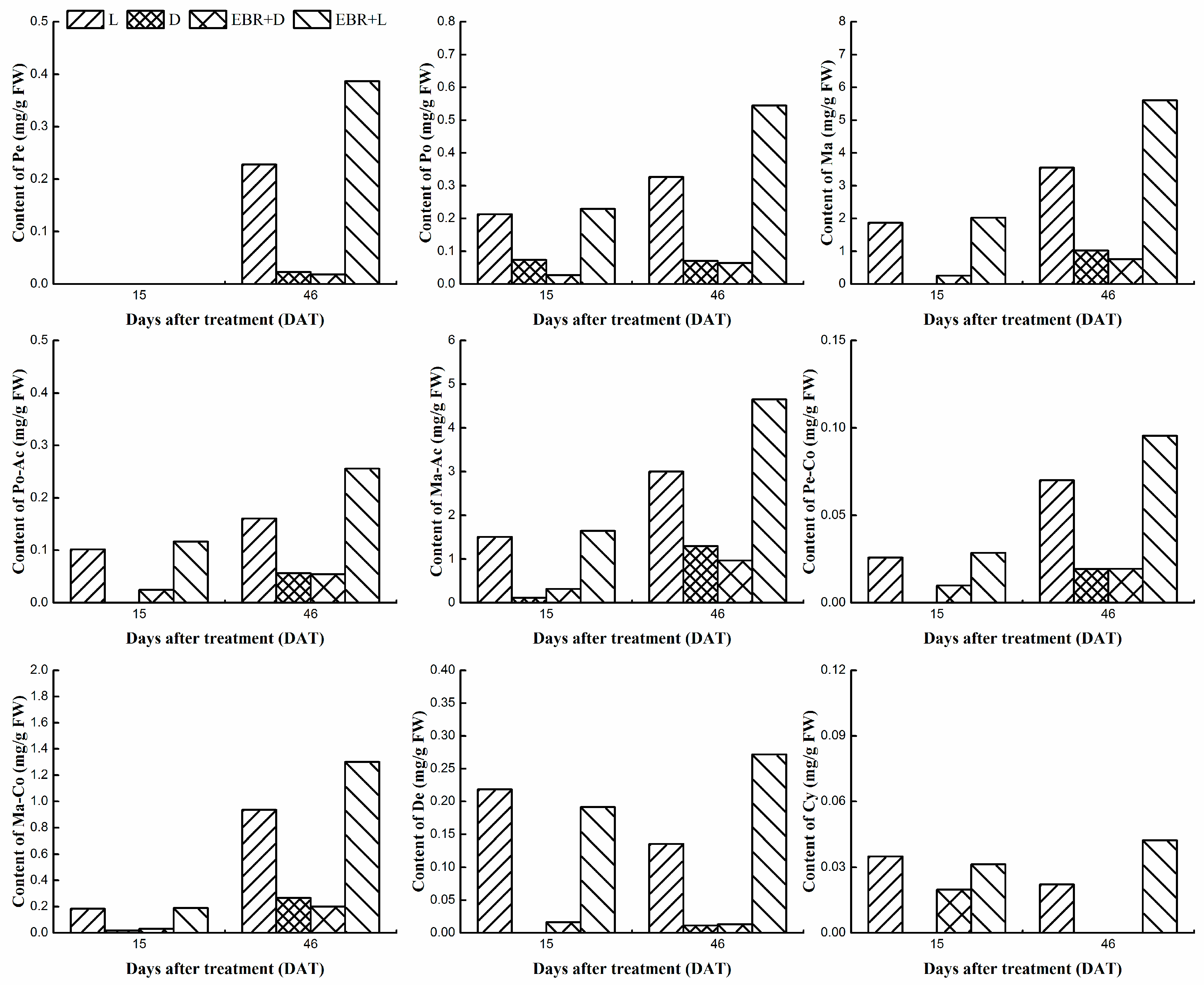
2.3. Determination Monomer Anthocyanins Content in Grape Skins
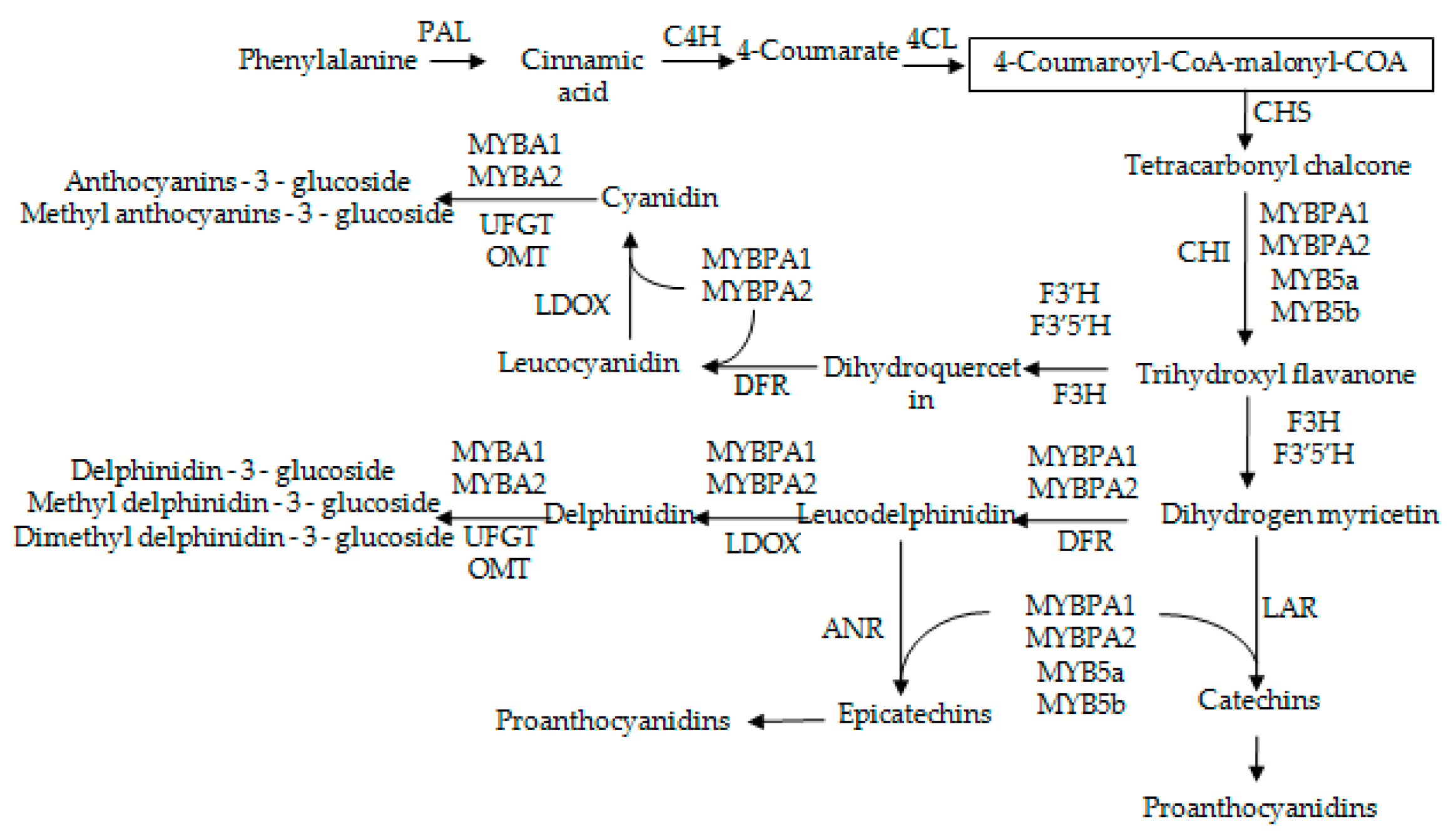
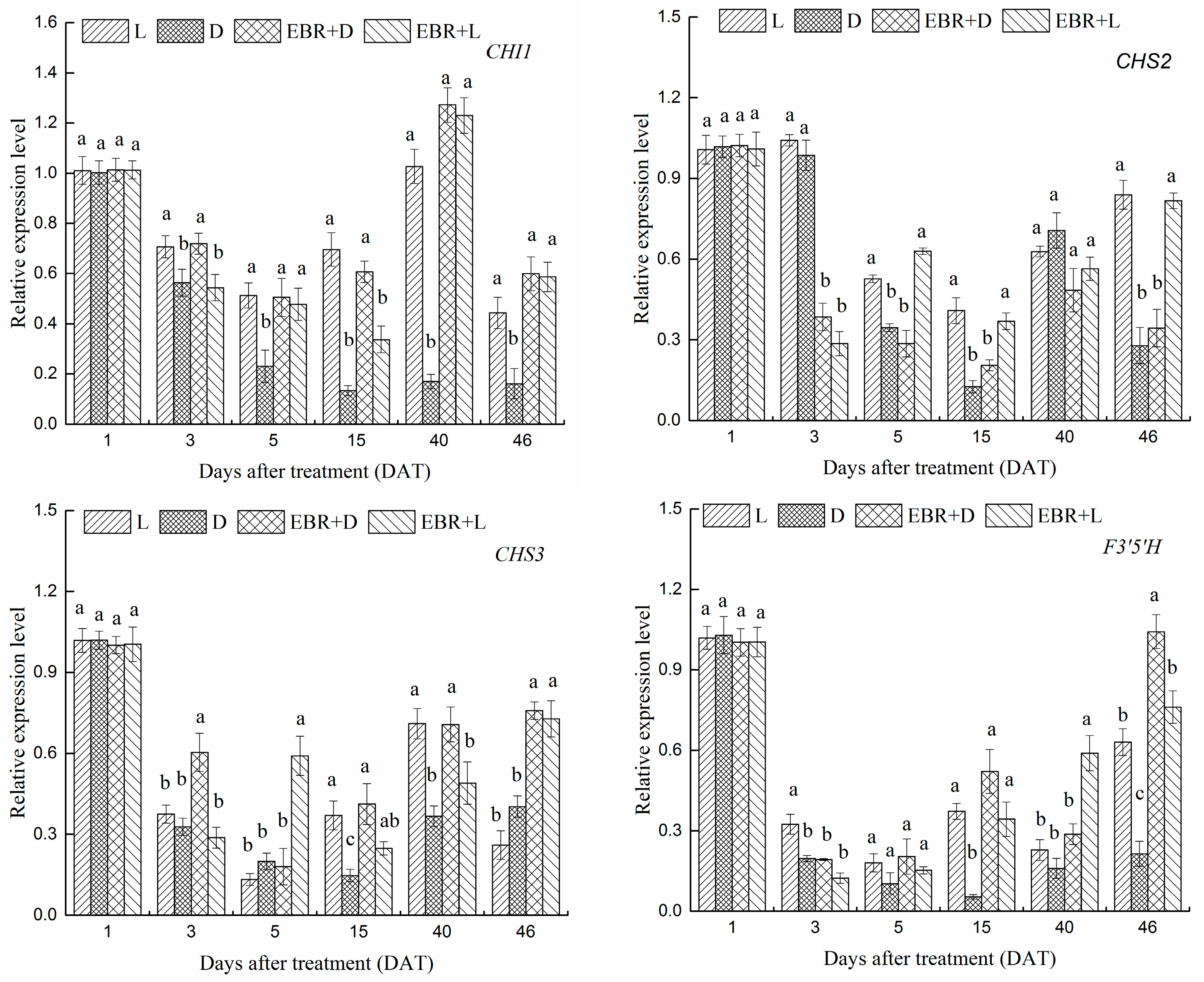
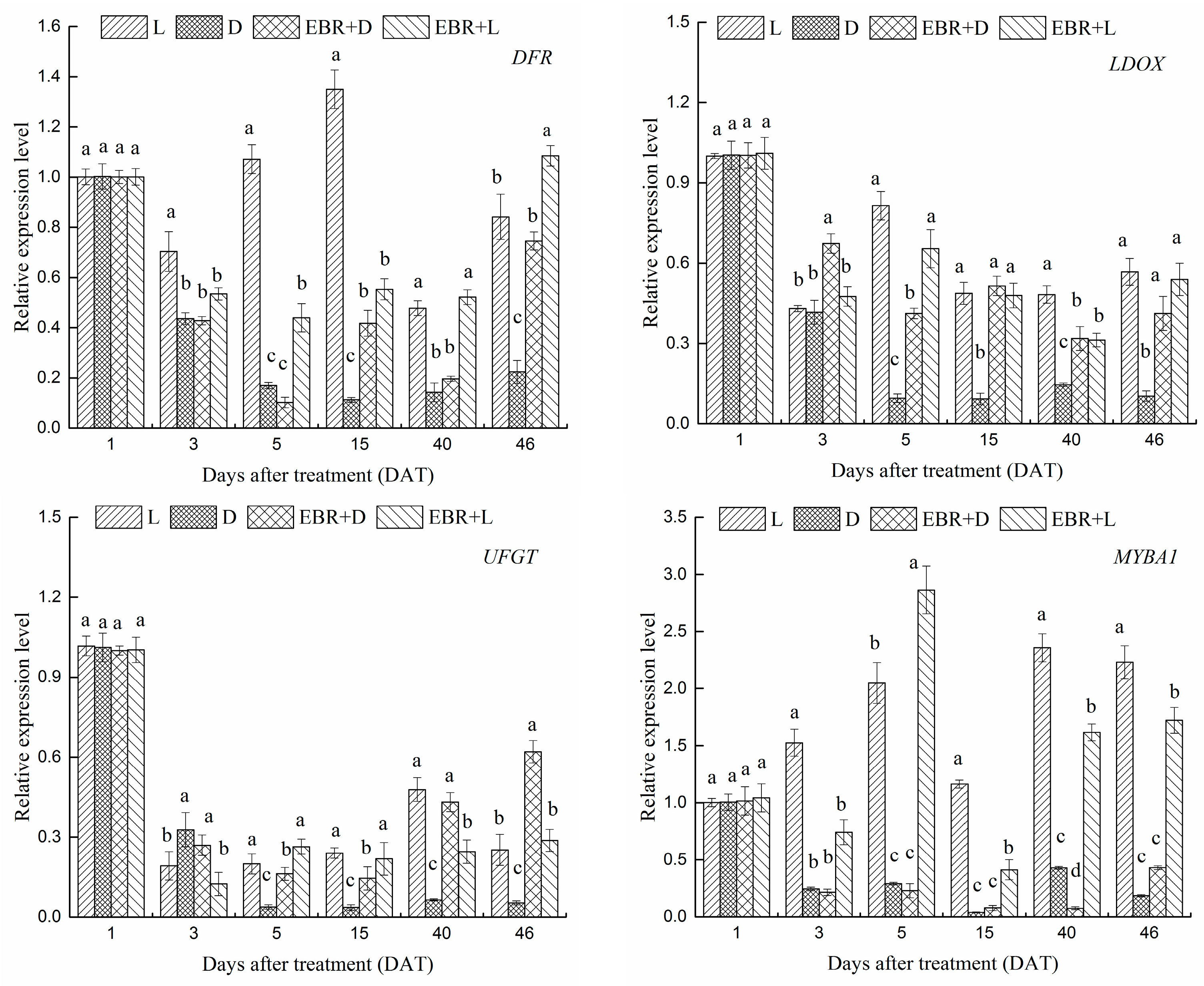
2.4. The Expression Patterns of Anthocyanins Biosynthetic Genes and Transcriptional Regulator Genes in Grape Skins
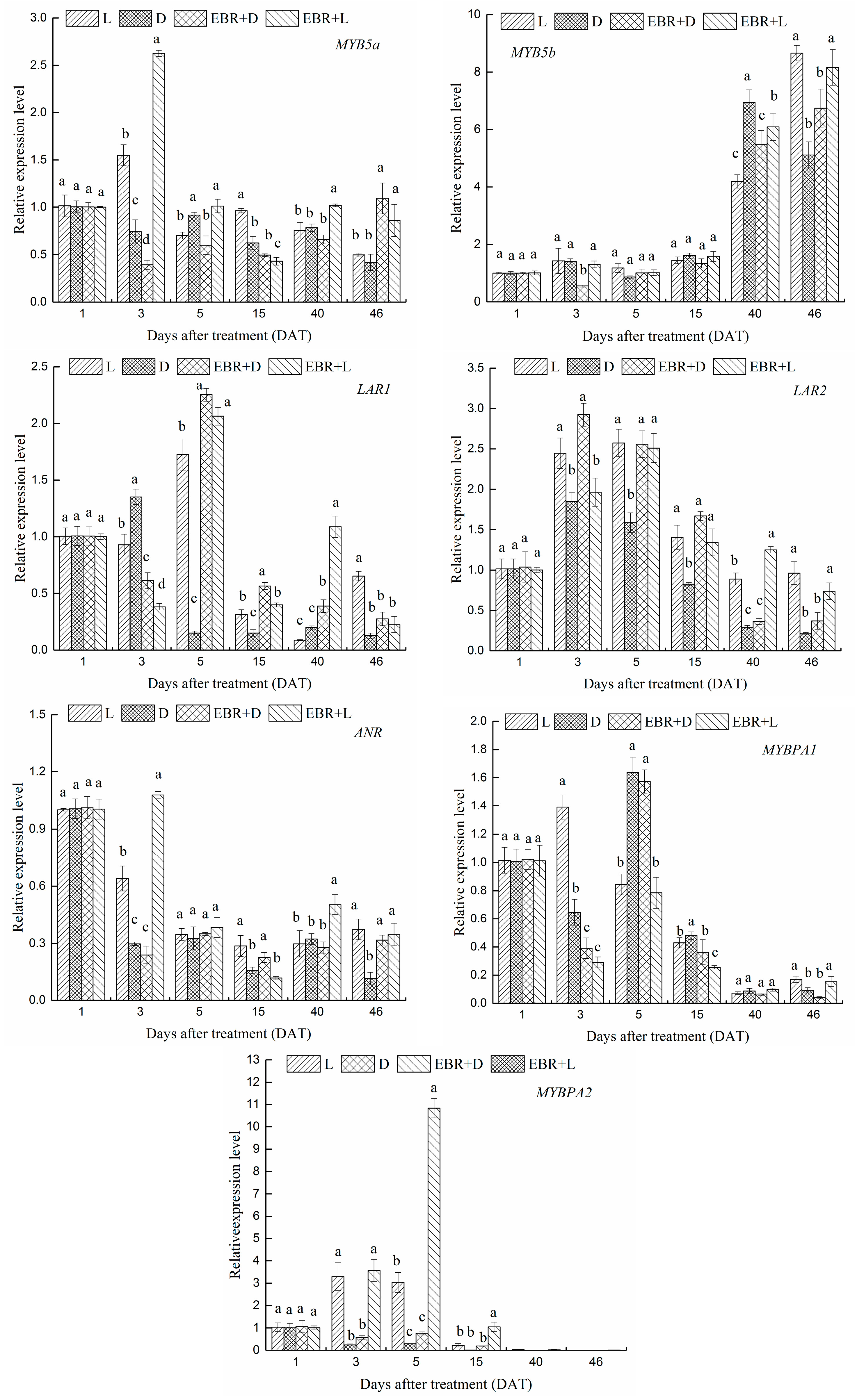
2.5. The Expression Patterns of Pas Biosynthetic Genes and Transcriptional Regulator Genes in Grape Skins
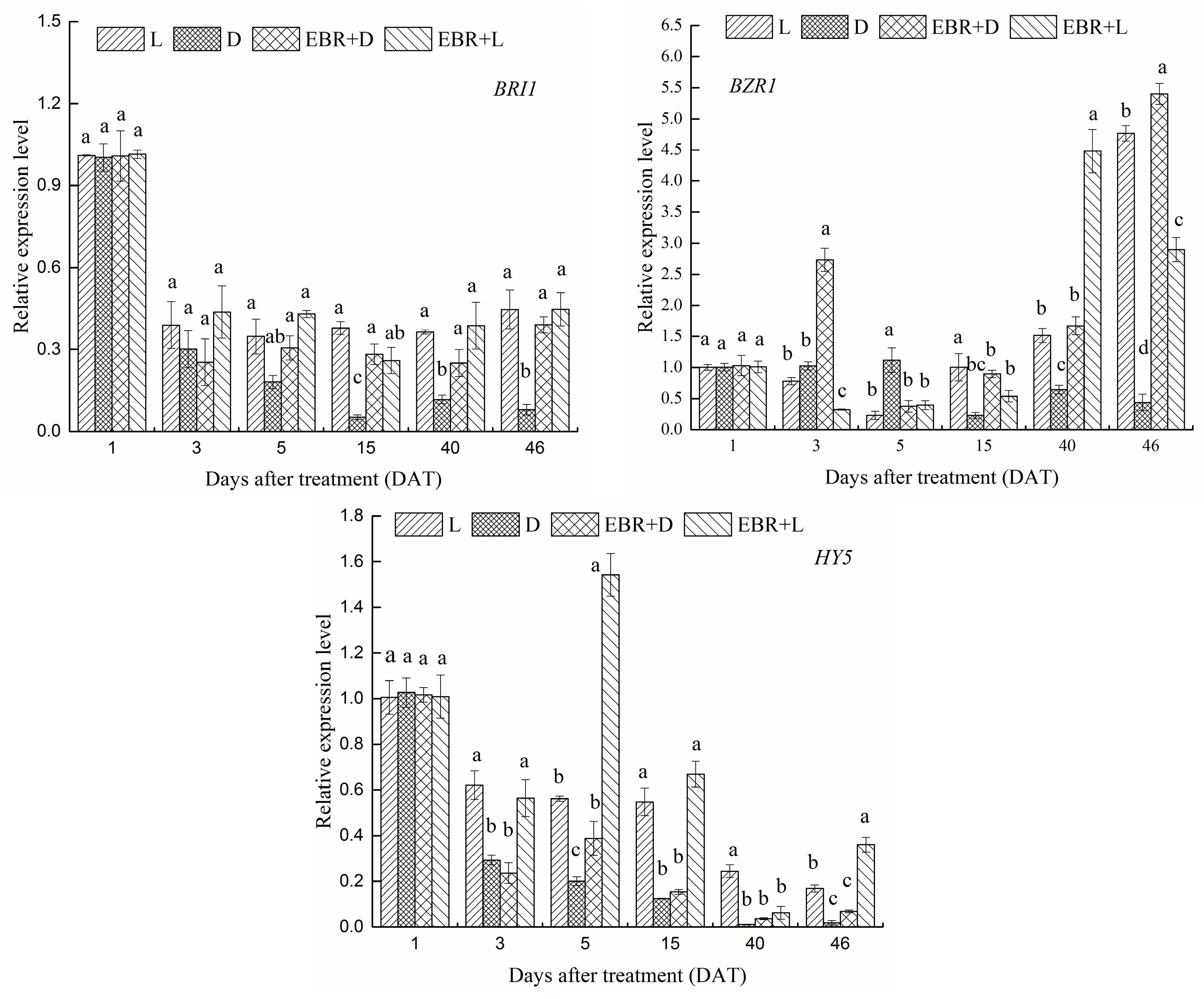
2.6. Expression Patterns of VvBZR1, VvHY5 and VvBRI1 Transcriptional Regulator Genes in Grape Skins
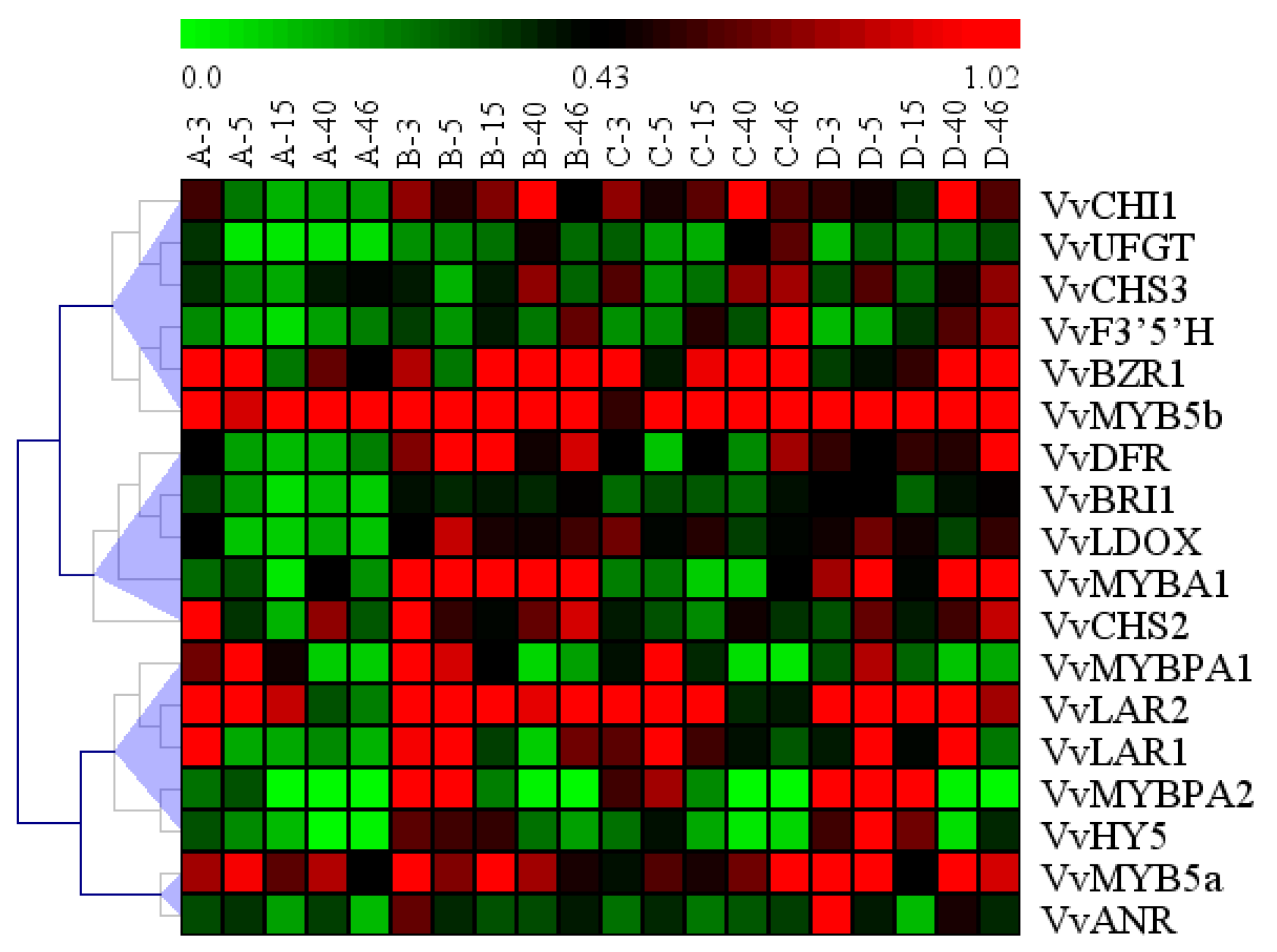
2.7. Hierarchical Clustering in the Profiles of All Genes Analyzed
3. Discussion
4. Materials and Methods
4.1. Sample Treatments
4.2. Experimental Design and Samples Collection
4.3. Determination the Physicochemical Indices of Berries
4.4. Extraction of Phenolic Compounds From Grape Skins
4.5. Determination of Phenolics Content
4.6. Determination the Content of Monomer Anthocyanins
4.7. RNA Extraction and cDNA Synthesis Real-Time Quantitative PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- González-Neves, G.; Gil, G.; Barreiro, L. Influence of grape variety on the extraction of anthocyanins during the fermentation on skins. Eur. Food Res. Technol. 2008, 226, 1349–1355. [Google Scholar] [CrossRef]
- Lorenzis, G.D.; Rustioni, L.; Parisi, S.G.; Zoli, F.; Brancadoro, L. Anthocyanin biosynthesis during berry development in corvina grape. Sci. Hortic. 2016, 212, 74–80. [Google Scholar] [CrossRef]
- Parpinello, G.P.; Versari, A.; Chinnici, F.; Galassi, S. Relationship among sensory descriptors, consumer preference and color parameters of Italian Novello red wines. Food Res. Int. 2010, 42, 1389–1395. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Loyola, R.; Herrera, D.; Mas, A.; Wong, D.C.; Höll, J.; Cavallini, E.; Amato, A.; Azuma, A.; Ziegler, T.; Aquea, F. The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 2016, 67, 5429–5445. [Google Scholar] [CrossRef] [PubMed]
- Shinomiya, R.; Fujishima, H.; Muramoto, K.; Shiraishi, M. Impact of temperature and sunlight on the skin coloration of the ‘Kyoho’ table grape. Sci. Hortic. 2015, 193, 77–83. [Google Scholar] [CrossRef]
- Wang, X.; Wei, Z.; Ma, F. The effects of fruit bagging on levels of phenolic compounds and expression by anthocyanin biosynthetic and regulatory genes in red-fleshed apples. Process Biochem. 2015, 50, 1774–1782. [Google Scholar] [CrossRef]
- Zhang, C.; Jia, H.; Wu, W.; Wang, X.; Fang, J.; Wang, C. Functional conservation analysis and expression modes of grape anthocyanin synthesis genes responsive to low temperature stress. Gene 2015, 574, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Winkelshirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Dai, Z.W.; Meddar, M.; Renaud, C.; Merlin, I.; Hilbert, G.; Delrot, S.; Gomès, E. Long-term in vitro culture of grape berries and its application to assess the effects of sugar supply on anthocyanin accumulation. J. Exp. Bot. 2014, 65, 4665–4677. [Google Scholar] [CrossRef] [PubMed]
- António, T.; José, E.D.; Castellarin, S.D.; Gerós, H. Berry Phenolics of Grapevine under Challenging Environments. Int. J. Mol. Sci. 2013, 14, 18711–18739. [Google Scholar] [CrossRef]
- Boss, P.K.; Davies, C.; Robinson, S.P. Anthocyanin composition and anthocyanin pathway gene expression in grapevine sports differing in berry skin colour. Aust. J. Grape Wine Res. 1996, 2, 163–170. [Google Scholar] [CrossRef]
- Costantini, L.; Malacarne, G.; Lorenzi, S.; Troggio, M.; Mattivi, F.; Moser, C.; Grando, M.S. New candidate genes for the fine regulation of the colour of grapes. J. Exp. Bot. 2015, 66, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Da, S.F.; Iandolino, A.; Al-Kayal, F.; Bohlmann, M.C.; Cushman, M.A.; Lim, H.; Ergul, A.; Figueroa, R.; Kabuloglu, E.K.; Osborne, C. Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiol. 2005, 139, 574–597. [Google Scholar]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef] [PubMed]
- Fournierlevel, A.; Hugueney, P.; Verriès, C.; This, P.; Ageorges, A. Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.). BMC Plant Biol. 2011, 11, 269–287. [Google Scholar]
- Ali, M.B.; Howard, S.; Chen, S.; Wang, Y.; Yu, O.; Kovacs, L.G.; Qiu, W. Berry skin development in Norton grape: Distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol. 2011, 11, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Zoratti, L.; Karppinen, K.; Luengo, E.A.; Häggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.Z.; Hu, F.C.; Hu, G.B.; Li, X.J.; Huang, X.M.; Wang, H.C. Differential Expression of Anthocyanin Biosynthetic Genes in Relation to Anthocyanin Accumulation in the Pericarp of Litchi Chinensis Sonn. PLoS ONE 2011, 6, e19455. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, A.; Fu, C.H.; Gui, B.H.; Zhi, C.Z. Functional analysis of the UDP glucose: Flavonoid-3-Oglucosyltransferase (UFGT) promoter from litchi (Litchi chinesis Sonn.) and transient expression in onions (Allium cepa Linn.). Afr. J. Plant Sci. 2015, 9, 244–249. [Google Scholar] [CrossRef]
- HE, F.; He, J.J.; Pan, Q.H.; Duan, C.Q. Mass-spectrometry evidence confirming the presence of pelargonidin-3-O-glucoside in the berry skins of Cabernet Sauvignon and Pinot Noir (Vitis vinifera L.). Aust. J. Grape Wine Res. 2010, 16, 464–468. [Google Scholar] [CrossRef]
- Meng, R.; Qu, D.; Liu, Y.; Gao, Z.; Yang, H.; Shi, X.; Zhao, Z. Anthocyanin accumulation and related gene family expression in the skin of dark-grown red and non-red apples (Malus domestica Borkh.) in response to sunlight. Sci. Hortic. 2015, 189, 66–73. [Google Scholar] [CrossRef]
- Zhao, Q.; He, F.; Reeves, M.J.; Pan, Q.H.; Duan, C.Q.; Wang, J. Expression of structural genes related to anthocyanin biosynthesis of Vitis amurensis. J. For. Res. 2016, 27, 647–657. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Gambetta, G.A.; Wada, H.; Krasnow, M.N.; Cramer, G.R.; Peterlunger, E.; Shackel, K.A.; Matthews, M.A. Characterization of major ripening events during softening in grape: Turgor, sugar accumulation, abscisic acid metabolism, colour development, and their relationship with growth. J. Exp. Bot. 2016, 67, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Song, C.Z.; Chi, M.; Wang, T.M.; Zuo, L.L.; Li, X.L.; Zhang, Z.W.; Xi, Z.M. The effects of light and ethylene and their interaction on the regulation of proanthocyanidin and anthocyanin synthesis in the skins of Vitis vinifera berries. Plant Growth Regul. 2016, 79, 1–14. [Google Scholar] [CrossRef]
- Gomes, M.M.A. Physiological effects related to brassinosteroid application in plants. In Brassinosteroids: A Class of Plant Hormone; Hayat, S., Ahmad, A., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 193–242. [Google Scholar]
- Xi, Z.M.; Zhang, Z.W.; Huo, S.S.; Luan, L.Y.; Gao, X.; Ma, L.N.; Fang, Y.L. Regulating the secondary metabolism in grape berry using exogenous 24-epibrassinolide for enhanced phenolics content and antioxidant capacity. Food Chem. 2013, 141, 3056–3065. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Ahmad, A. Brassinosteroids: A Class of Plant Hormone; Springer: Dordrecht, The Netherlands, 2011; pp. 1283–1317. [Google Scholar]
- Hayat, S.; Ali, B.; Hasan, S.A.; Ahmad, A. Brassinosteroid enhanced the level of antioxidants under cadmium stress in Brassica juncea. Environ. Exp. Bot. 2007, 60, 33–41. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, P.; Meng, J.; Xi, Z. Effect of exogenous 24-epibrassinolide on chlorophyll fluorescence, leaf surface morphology and cellular ultrastructure of grape seedlings (Vitis vinifera L.) under water stress. Acta Physiol. Plant. 2015, 37, 1–12. [Google Scholar] [CrossRef]
- Cui, L.; Zou, Z.; Zhang, J.; Zhao, Y.; Yan, F. 24-Epibrassinoslide enhances plant tolerance to stress from low temperatures and poor light intensities in tomato (Lycopersicon esculentum Mill.). Funct. Integr. Genom. 2016, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Asghari, M.; Zahedipour, P. 24-Epibrassinolide Acts as a Growth-Promoting and Resistance-Mediating Factor in Strawberry Plants. J. Plant Growth Regul. 2016, 35, 1–8. [Google Scholar] [CrossRef]
- Liu, Q.; Xi, Z.; Gao, J.; Meng, Y.; Lin, S.; Zhang, Z. Effects of exogenous 24-epibrassinolide to control grey mould and maintain postharvest quality of table grapes. Int. J. Food Sci. Technol. 2016, 51, 1236–1243. [Google Scholar] [CrossRef]
- Xu, F.; Gao, X.; Xi, Z.M.; Zhang, H.; Peng, X.Q.; Wang, Z.Z.; Wang, T.M.; Meng, Y. Application of exogenous 24-epibrassinolide enhances proanthocyanidin biosynthesis in Vitis vinifera ‘Cabernet Sauvignon’ berry skin. Plant Growth Regul. 2015, 75, 741–750. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid Signaling Network and Regulation of Photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Lee, J. Light exclusion influence on grape anthocyanin. Heliyon 2017, 3, e00243. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, E.; Choi, G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Song, Z.; Zhang, H. Repression of MYBL2 by both microRNA858a and HY5 Leads to the Activation of Anthocyanin Biosynthetic Pathway in Arabidopsis. Mol. Plant 2016, 9, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Bureau, S.M.; Baumes, R.L.; Razungles, A.J. Effects of vine or bunch shading on the glycosylated flavor precursors in grapes of Vitis vinifera L. Cv. syrah. J. Agric. Food Chem. 2000, 48, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Friedel, M. Impact of light exposure on fruit composition of white ‘Riesling’ grape berries (“Vitis vinifera” L.). J. Grapevine Res. 2015, 54, 107–116. [Google Scholar]
- Sharma, R.R.; Reddy, S.V.R.; Jhalegar, M.J. Pre-harvest fruit bagging: A useful approach for plant protection and improved post-harvest fruit quality—A review. J. Hortic. Sci. Biotechnol. 2014, 89, 101–113. [Google Scholar] [CrossRef]
- Lorrain, B.; Chira, K.; Teissedre, P.L. Phenolic composition of Merlot and Cabernet-Sauvignon grapes from Bordeaux vineyard for the 2009-vintage: Comparison to 2006, 2007 and 2008 vintages. Food Chem. 2011, 126, 1991–1999. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Ortín, A.B.; Rodríguez-Rodríguez, P.; Gil-Muñoz, R.; Jiménez-Pascual, E.; Busse-Valverde, N.; Martínez-Cutillas, A.; López-Roca, J.M.; Gómez-Plaza, E. Influence of berry ripeness on concentration, qualitative composition and extractability of grape seed tannins. Aust. J. Grape Wine Res. 2012, 18, 123–130. [Google Scholar] [CrossRef]
- Bucchetti, B.; Matthews, M.A.; Falginella, L.; Peterlunger, E.; Castellarin, S.D. Effect of water deficit on Merlot grape tannins and anthocyanins across four seasons. Sci. Hortic. 2011, 128, 297–305. [Google Scholar] [CrossRef]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin Synthesis and Expression of Genes Encoding Leucoanthocyanidin Reductase and Anthocyanidin Reductase in Developing Grape Berries and Grapevine Leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Gagné, S.; Lacampagne, S.; Claisse, O.; Gény, L. Leucoanthocyanidin reductase and anthocyanidin reductase gene expression and activity in flowers, young berries and skins of Vitis vinifera L. cv. Cabernet-Sauvignon during development. Plant Physiol. Biochem. 2009, 47, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.F.; Ji, W.; Gao, M.Y.; Niu, T.Q.; Xing, Y.F.; Niu, X.Y. Accumulation of flavanols and expression of leucoanthocyanidin reductase induced by postharvest UV-C irradiation in grape berry. Genet. Mol. Res. 2015, 14, 7687–7695. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.L.; Liu, M.; Zhao, H.; Meng, J.F.; Fang, Y.L. Effect of Exogenous Abscisic Acid and Methyl Jasmonate on Anthocyanin Composition, Fatty Acids, and Volatile Compounds of Cabernet Sauvignon (Vitis vinifera L.). Grape Berries. Molecules 2016, 21, E1354. [Google Scholar] [CrossRef] [PubMed]
- Luan, L.Y.; Zhang, Z.W.; Xi, Z.M.; Huo, S.S.; Ma, L.N. Comparing the effects of exogenous abscisic acid on the phenolic composition of Yan 73 and Cabernet Sauvignon (Vitis vinifera L.) wines. Eur. Food Res. Technol. 2014, 239, 203–213. [Google Scholar] [CrossRef]
- Villalobos-González, L.; Peña-Neira, A.; Ibáñez, F.; Pastenes, C. Long-term effects of abscisic acid (ABA) on the grape berry phenylpropanoid pathway: Gene expression and metabolite content. Plant Physiol. Biochem. 2016, 105, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Peng, Q.Z.; Du, C.; Li, K.G.; Xie, D.Y. Characterization of Flavan-3-ols and Expression of MYB and Late Pathway Genes Involved in Proanthocyanidin Biosynthesis in Foliage of Vitis bellula. Metabolites 2013, 3, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Bogs, J.; Jaffé, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef] [PubMed]
- Terrier, N.; Torregrosa, L.; Ageorges, A.; Vialet, S.; Verries, C.; Cheynier, V.; Romieu, C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar] [CrossRef] [PubMed]
- Koyama, K.; Numata, M.; Nakajima, I.; Goto-Yamamoto, N.; Matsumura, H.; Tanaka, N. Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J. Exp. Bot. 2014, 65, 4433–4449. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Deng, Z.; Wang, Z.Y. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr. Opin. Plant Biol. 2009, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Li, J. Regulation of the Nuclear Activities of Brassinosteroid Signaling. Curr. Opin. Plant Biol. 2010, 13, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Azuma, A.; Fujii, H.; Shimada, T.; Yakushiji, H. Microarray Analysis for the Screening of Genes Inducible by Light or Low Temperature in Post-veraison Grape Berries. Hortic. J. 2015, 84, 214–216. [Google Scholar] [CrossRef]
- Coombe, B.G. Growth Stages of the Grapevine: Adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 2008, 1, 104–110. [Google Scholar] [CrossRef]
- Stefano, R.D.; Cravero, M.C. The grape phenolic determination. Riv. Vitic. Enol. 1991, xliv(58), 37–45. [Google Scholar]
- Harbertson, J.F.; Picciotto, E.A.; Adams, D.O. Measurement of polymeric pigments in grape berry extracts and wines using a protein precipitation assay combined with bisulfite bleaching. Am. J. Enol. Vitic. 2003, 54, 301–306. [Google Scholar]
- Jayaprakasha, G.K.; Singh, R.P.; Sakariah, K.K. Antioxidant activity of grape seed (Vitis vinifera) extracts on peroxidation models in vitro. Food Chem. 2001, 73, 285–290. [Google Scholar] [CrossRef]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Yuan, C.; Ruan, S.; Zhang, Z.; Meng, J.; Xi, Z. Exogenous 24-Epibrassinolide Interacts with Light to Regulate Anthocyanin and Proanthocyanidin Biosynthesis in Cabernet Sauvignon (Vitis vinifera L.). Molecules 2018, 23, 93. https://doi.org/10.3390/molecules23010093
Zhou Y, Yuan C, Ruan S, Zhang Z, Meng J, Xi Z. Exogenous 24-Epibrassinolide Interacts with Light to Regulate Anthocyanin and Proanthocyanidin Biosynthesis in Cabernet Sauvignon (Vitis vinifera L.). Molecules. 2018; 23(1):93. https://doi.org/10.3390/molecules23010093
Chicago/Turabian StyleZhou, Yali, Chunlong Yuan, Shicheng Ruan, Zhenwen Zhang, Jiangfei Meng, and Zhumei Xi. 2018. "Exogenous 24-Epibrassinolide Interacts with Light to Regulate Anthocyanin and Proanthocyanidin Biosynthesis in Cabernet Sauvignon (Vitis vinifera L.)" Molecules 23, no. 1: 93. https://doi.org/10.3390/molecules23010093



