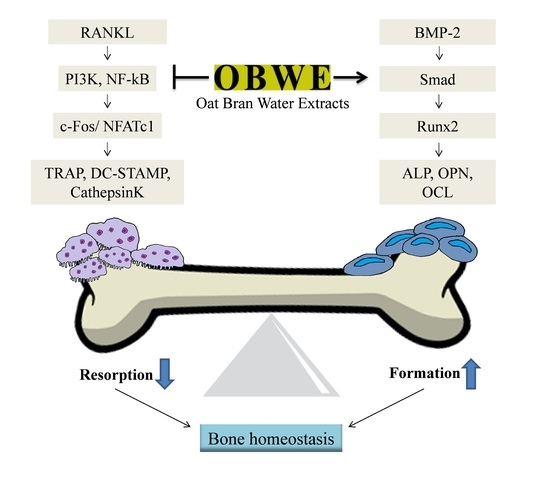
The Dual Role of Oat Bran Water Extract in Bone Homeostasis Through the Regulation of Osteoclastogenesis and Osteoblast Differentiation
Abstract
:1. Introduction
2. Results and Discussion
2.1. OBWE Inhibits RANKL-Induced Osteoclast Differentiation
2.2. OBWE Inhibits RANKL-Related Expression of c-Fos and NFATc1 through the Modulation of NF-κB/I-κB Signaling Molecules
2.3. OBWE Enhances BMP-2-Mediated Osteoblast Differentiation in C2C12 Cells
2.4. OBWE Contributes to the BMP-2-Stimulated Expression of Runx2 through the Activation of Smad Signaling Pathways
2.5. OBWE Prevents Ovariectomy-Induced Bone Loss In Vivo
3. Materials and Methods
3.1. Preparation of the Oat Bran Water Extract
3.2. Reagents and Antibodies
3.3. Preparations of Osteoclast Precursor Cells
3.4. Osteoclast Cell Culture and Osteoclast Differentiation
3.5. TRAP Staining and Activity Assay
3.6. Cell Viability Assay
3.7. RNA Isolation and Real-Time Polymerase Chain Reaction Analysis
3.8. Western Blot Analysis
3.9. Osteoblast Differentiation
3.10. Alkaline Phosphatase Staining and Activity Assays
3.11. Ovariectomy-Induced Bone Erosion
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klibanski, A.; Adams-Campbell, L.; Bassford, T.; Blair, S.N.; Boden, S.D.; Dickersin, K.; Gifford, D.R.; Glasse, L.; Goldring, S.R.; Hruska, K.; et al. Osteoporosis prevention, diagnosis, and therapy. JAMA-J. Am. Med. Assoc. 2001, 285, 785–795. [Google Scholar]
- Karsenty, G.; Wagner, E.F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2002, 2, 389–406. [Google Scholar] [CrossRef]
- Rodan, G.A.; Martin, T.J. Therapeutic approaches to bone diseases. Science 2000, 289, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Khosla, S.; Riggs, B.L. Pathophysiology of age-related bone loss and osteoporosis. Endocrin. Metab. Clin. 2005, 34, 1015–1030. [Google Scholar] [CrossRef] [PubMed]
- Manolagas, S.C.; Parfitt, A.M. What old means to bone. Trends Endocrin. Met. 2010, 21, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Lee, Z.H.; Kim, H.H. Signal transduction by receptor activator of nuclear factor kappa B in osteoclasts. Biochem. Bioph. Res. Commun. 2003, 305, 211–214. [Google Scholar] [CrossRef]
- Feng, X. RANKing intracellular signaling in osteoclasts. Iubmb Life 2005, 57, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 2007, 7, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H. Inflammatory bone destruction and osteoimmunology. J. Periodontal Res. 2005, 40, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Grigoriadis, A.E.; Wang, Z.Q.; Cecchini, M.G.; Hofstetter, W.; Felix, R.; Fleisch, H.A.; Wagner, E.F. C-Fos—A Key Regulator of Osteoclast-Macrophage Lineage Determination and Bone Remodeling. Science 1994, 266, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H.; Kim, S.; Koga, T.; Nishina, H.; Isshiki, M.; Yoshida, H.; Saiura, A.; Isobe, M.; Yokochi, T.; Inoue, J.; et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 2002, 3, 889–901. [Google Scholar] [CrossRef]
- Teitelbaum, S.L. Bone resorption by osteoclasts. Science 2000, 289, 1504–1508. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almalki, S.G.; Agrawal, D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 2016, 92, 41–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollinger, J.O.; Schmitt, J.M.; Buck, D.C.; Shannon, R.; Joh, S.P.; Zegzula, H.D.; Wozney, J. Recombinant human bone morphogenetic protein-2 and collagen for bone regeneration. J. Biomed. Mater. Res. 1998, 43, 356–364. [Google Scholar] [CrossRef]
- Cheng, H.W.; Jiang, W.; Phillips, F.M.; Haydon, R.C.; Peng, Y.; Zhou, L.; Luu, H.H.; An, N.L.; Breyer, B.; Vanichakarn, P.; et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J. Bone Jt. Surg. Am. 2003, 85-A, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Herpin, A.; Cunningham, C. Cross-talk between the bone morphogenetic protein pathway and other major signaling pathways results in tightly regulated cell-specific outcomes. Febs J. 2007, 274, 2977–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, J.B.; Stein, G.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Montecino, M.; Hassan, M.Q.; Gaur, T.; Lengner, C.J.; Young, D.W. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Dis. 2006, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kahn, H.S.; Tatham, L.M.; Rodriguez, C.; Calle, E.E.; Thun, M.J.; Heath, C.W. Stable behaviors associated with adults’ 10-year change in body mass index and likelihood of gain at the waist. Am. J. Public Health 1997, 87, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Bruderer, M.; Richards, R.G.; Alini, M.; Stoddart, M.J. Role and Regulation of Runx2 in Osteogenesis. Eur. Cells Mater. 2014, 28, 269–286. [Google Scholar] [CrossRef]
- Sultan, M.T.; Butt, M.S.; Qayyum, M.M.; Suleria, H.A. Immunity: plants as effective mediators. Crit. Rev. Food Sci. 2014, 54, 1298–1308. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; Saeed, F.; Anjum, F.M.; Afzaal, M.; Tufail, T.; Bashir, M.S.; Ishtiaq, A.; Hussain, S.; Suleria, H.A. Natural polyphenols: An overview. Int. J. Food Prop. 2017, 20, 1689–1699. [Google Scholar] [CrossRef]
- Bryngelsson, S.; Mannerstedt-Fogelfors, B.; Kamal-Eldin, A.; Andersson, R.; Dimberg, L.H. Lipids and antioxidants in groats and hulls of Swedish oats (Avena sativa L). J. Sci. Food Agr. 2002, 82, 606–614. [Google Scholar] [CrossRef]
- Dimberg, L.H.; Gissen, C.; Nilsson, J. Phenolic compounds in oat grains (Avena sativa L.) grown in conventional and organic systems. Ambio 2005, 34, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Emmons, C.L.; Peterson, D.M.; Paul, G.L. Antioxidant capacity of oat (Avena sativa L.) extracts. 2. In vitro antioxidant activity and contents of phenolic and tocol antioxidants. J. Agr. Food Chem. 1999, 47, 4894–4898. [Google Scholar] [CrossRef]
- Bahraminejad, S.; Asenstorferi, R.E.; Rileyi, I.T.; Schultz, C.J. Analysis of the antimicrobial activity of flavonoids and saponins isolated from the shoots of oats (Avena sativa L.). J. Phytopatho. 2008, 156, 1–7. [Google Scholar] [CrossRef]
- Tapola, N.; Karvonen, H.; Niskanen, L.; Mikola, M.; Sarkkinen, E. Glycemic responses of oat bran products in type 2 diabetic patients. Nutr. Metab. Cardiovas. 2005, 15, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.M.; Nie, L.; Wu, D.Y.; Wise, M.L.; Collins, F.W.; Meydani, S.N.; Meydani, M. Avenanthramides Inhibit Proliferation of Human Colon Cancer Cell Lines In Vitro. Nutr. Cancer 2010, 62, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Ozkaya, H.; Ozkaya, B.; Duman, B.; Turksoy, S. Effect of Dephytinization by Fermentation and Hydrothermal Autoclaving Treatments on the Antioxidant Activity, Dietary Fiber, and Phenolic Content of Oat Bran. J. Agr. Food Chem. 2017, 65, 5713–5719. [Google Scholar] [CrossRef] [PubMed]
- Robitaille, J.; Fontaine-Bisson, B.; Couture, P.; Tchernof, A.; Vohl, M.C. Effect of an oat bran-rich supplement on the metabolic profile of overweight premenopausal women. Ann. Nut. Metab. 2005, 49, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Varga, N.; Vereb, Z.; Rajnavolgyi, E.; Nemet, K.; Uher, F.; Sarkadi, B.; Apati, A. Mesenchymal stem cell like (MSCl) cells generated from human embryonic stem cells support pluripotent cell growth. Biochem. Bioph. Res. Commun. 2011, 414, 474–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raisz, L.G. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J. Clin. Investig. 2005, 115, 3318–3325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asagiri, M.; Takayanagi, H. The molecular understanding of osteoclast differentiation. Bone 2007, 40, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Ishida, N.; Hayashi, K.; Hoshijima, M.; Ogawa, T.; Koga, S.; Miyatake, Y.; Kumegawa, M.; Kimura, T.; Takeya, T. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J. Biol. Chem. 2002, 277, 41147–41156. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Ovitt, C.; Grigoriadis, A.E.; Mohlesteinlein, U.; Ruther, U.; Wagner, E.F. Bone and Hematopoietic Defects in Mice Lacking C-Fos. Nature 1992, 360, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.C.; Robinson, L.J.; Zaidi, M. Osteoclast signalling pathways. Biochem. Bioph. Res. Commun. 2005, 328, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Kadono, Y.; Naito, A.; Matsumoto, K.; Yamamoto, T.; Tanaka, S.; Inoue, J. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. Embo J. 2001, 20, 1271–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iotsova, V.; Caamano, J.; Loy, J.; Yang, Y.; Lewin, A.; Bravo, R. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nat. Med. 1997, 3, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.D.; Harrison, S.C. Structure of an IkappaBalpha/NF-kappaB complex. Cell 1998, 95, 749–758. [Google Scholar] [CrossRef]
- Jang, W.G.; Kim, E.J.; Kim, D.K.; Ryoo, H.M.; Lee, K.B.; Kim, S.H.; Choi, H.S.; Koh, J.T. BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J. Biol. Chem. 2012, 287, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Gersbach, C.A.; Byers, B.A.; Pavlath, G.K.; Garcia, A.J. Runx2/Cbfa1 stimulates transdifferentiation of primary skeletal myoblasts into a mineralizing osteoblastic phenotype. Exp. Cell Res. 2004, 300, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Awad, H.A.; Liu, S.; Mahlios, J.; Zhang, S.; Guilak, F.; Mayo, M.S.; Quarles, L.D. Selective Runx2-II deficiency leads to low-turnover osteopenia in adult mice. Dev. Biol. 2005, 283, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Harada, H.; Tagashira, S.; Fujiwara, M.; Ogawa, S.; Katsumata, T.; Yamaguchi, A.; Komori, T.; Nakatsuka, M. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J. Biol. Chem. 1999, 274, 6972–6978. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Deng, C.; Li, Y.P. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012, 8, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone morphogenetic proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.H.; Derynck, R. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell. Dev. Biol. 2005, 21, 659–693. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Harris, M.A.; Rossini, G.; Dunstan, C.R.; Dallas, S.L.; Feng, J.Q.; Mundy, G.R.; Harris, S.E. Bone morphogenetic protein 2 (BMP-2) enhances BMP-3, BMP-4, and bone cell differentiation marker gene expression during the induction of mineralized bone matrix formation in cultures of fetal rat calvarial osteoblasts. Calcif. Tissue Int. 1997, 60, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Li, H.; Sasaki, T.; Holman, D.; Beres, B.; Dumont, R.J.; Pittman, D.D.; Hankins, G.R.; Helm, G.A. Osteogenic potential of five different recombinant human bone morphogenetic protein adenoviral vectors in the rat. Gene Ther. 2003, 10, 1735–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.W.; Kim, S.H.; Lee, K.S.; Kang, H.J.; Lee, M.J.; Park, K.I.; Lee, J.H.; Park, K.D.; Seo, W.D. Barley. Seedling Extracts Inhibit RANKL-Induced Differentiation, Fusion, and Maturation of Osteoclasts in the Early-to-Late Stages of Osteoclastogenesis. Evid. Based Complement. Alternat. Med. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Lee, K.S.; Lee, J.H.; Kang, H.J.; Lee, M.J.; Kim, H.Y.; Park, K.I.; Kim, S.L.; Shin, H.K.; Seo, W.D. Suppression of Akt-HIF-1alpha signaling axis by diacetyl atractylodiol inhibits hypoxia-induced angiogenesis. BMB Rep. 2016, 49, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.W.; Choi, S.W.; Kim, H.J.; Lee, K.S.; Kim, S.H.; Kim, S.L.; Do, S.H.; Seo, W.D. Germinated soy germ with increased soyasaponin Ab improves BMP-2-induced bone formation and protects against in vivo bone loss in osteoporosis. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |






| Target Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| c-Fos | CCAGTCAAGAGCATCAGCAA | AAGTAGTGCAGCCCGGAGTA |
| NFATc1 | GGGTCAGTGTGACCGAAGAT | GGAAGTCAGAAGTGGGTGGA |
| TRAP | GATGACTTTGCCAGTCAGCA | ACATAGCCCACACCGTTCTC |
| OSCAR | AGGGAAACCTCATCCGTTTG | GAGCCGGAAATAAGGCACAG |
| DC-STAMP | CCAAGGAGTCGTCCATGATT | GGCTGCTTTGATCGTTTCTC |
| Cathepsin K | GGCCAACTCAAGAAGAAAAC | GTGCTTGCTTCCCTTCTGG |
| Runx2 | GACTGTGGTTACCGTCATGGC | ACTTGGTTTTTCATAACAGCGGA |
| ALP | GATGGCGTATGCCTCCTGCA | CGGTGGTGGGCCACAAAAGG |
| OPN | ACACTTTCACTCCAATCGTCC | TGCCCTTTCCGTTGTTGTCC |
| OCL | AGGGAAACCTCATCCGTTG | GAGCCGGAAATAAGGCACAG |
| GAPDH | ACCACAGTCCATGCCATCAC | TCCACCACCCTGTTGCTGTA |
| HPRT1 | TGCTCGAGATGTCATGAAGG | AGAGGTCCTTTTCACCAGCA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-H.; Kim, K.-J.; Kang, H.J.; Son, Y.-J.; Choi, S.-W.; Lee, M.-J. The Dual Role of Oat Bran Water Extract in Bone Homeostasis Through the Regulation of Osteoclastogenesis and Osteoblast Differentiation. Molecules 2018, 23, 3119. https://doi.org/10.3390/molecules23123119
Kim S-H, Kim K-J, Kang HJ, Son Y-J, Choi S-W, Lee M-J. The Dual Role of Oat Bran Water Extract in Bone Homeostasis Through the Regulation of Osteoclastogenesis and Osteoblast Differentiation. Molecules. 2018; 23(12):3119. https://doi.org/10.3390/molecules23123119
Chicago/Turabian StyleKim, Shin-Hye, Kwang-Jin Kim, Hyeon Jung Kang, Young-Jin Son, Sik-Won Choi, and Mi-Ja Lee. 2018. "The Dual Role of Oat Bran Water Extract in Bone Homeostasis Through the Regulation of Osteoclastogenesis and Osteoblast Differentiation" Molecules 23, no. 12: 3119. https://doi.org/10.3390/molecules23123119