Rapid Characterization and Identification of Non-Diterpenoid Constituents in Tinospora sinensis by HPLC-LTQ-Orbitrap MSn
Abstract
:1. Introduction
2. Results and Discussion
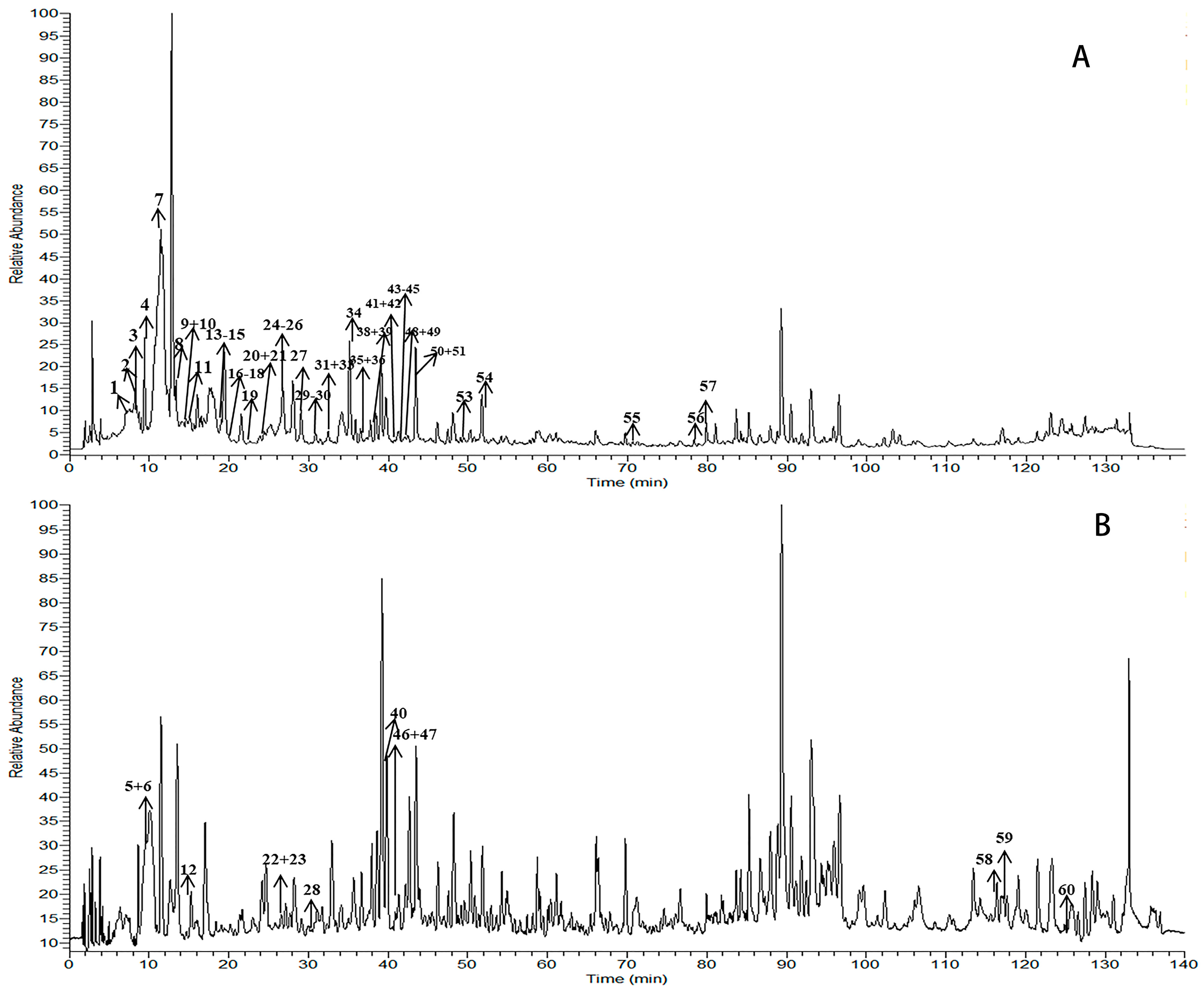
2.1. Identification of the Constituents by HPLC-LTQ-Orbitrap MSn
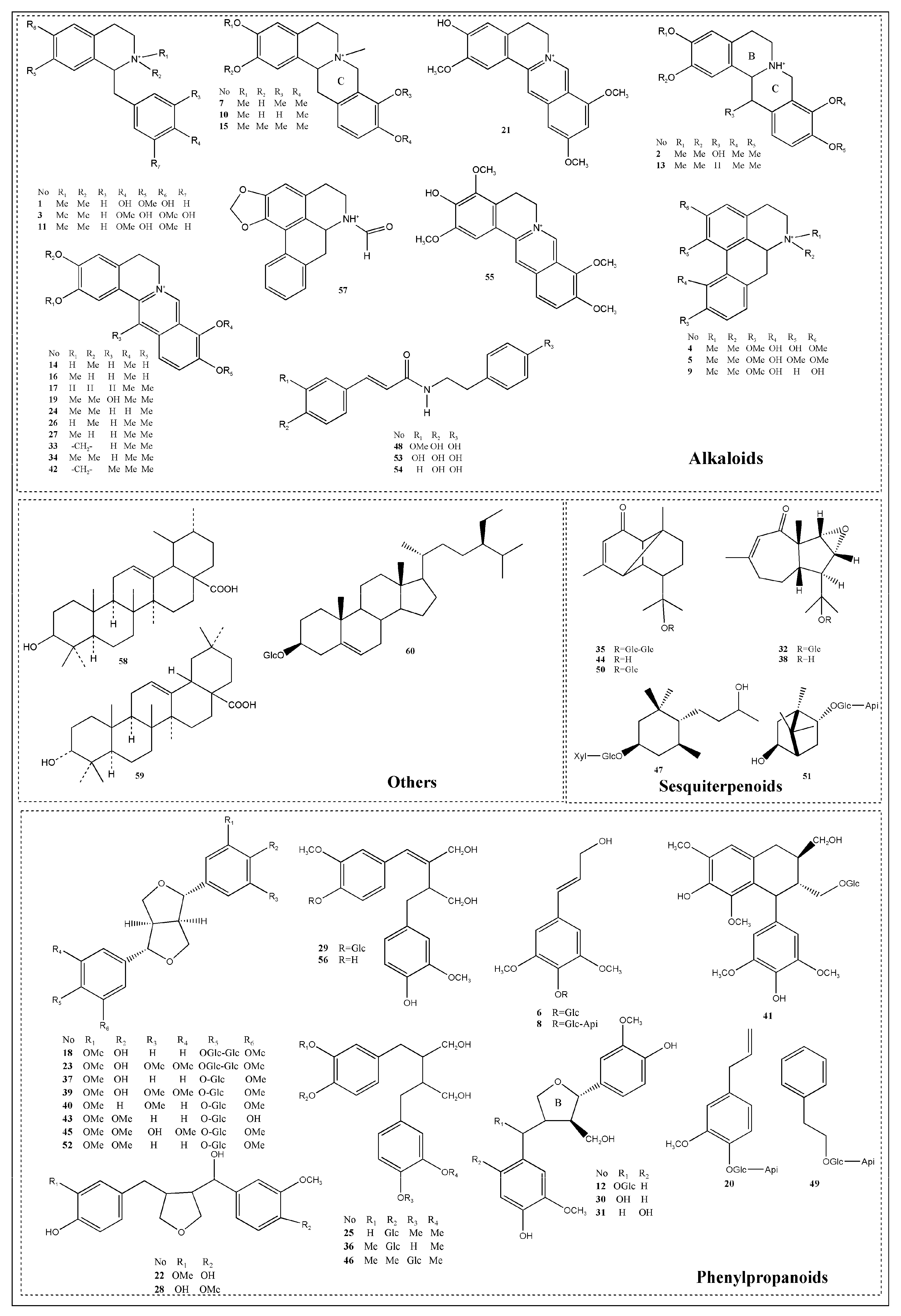
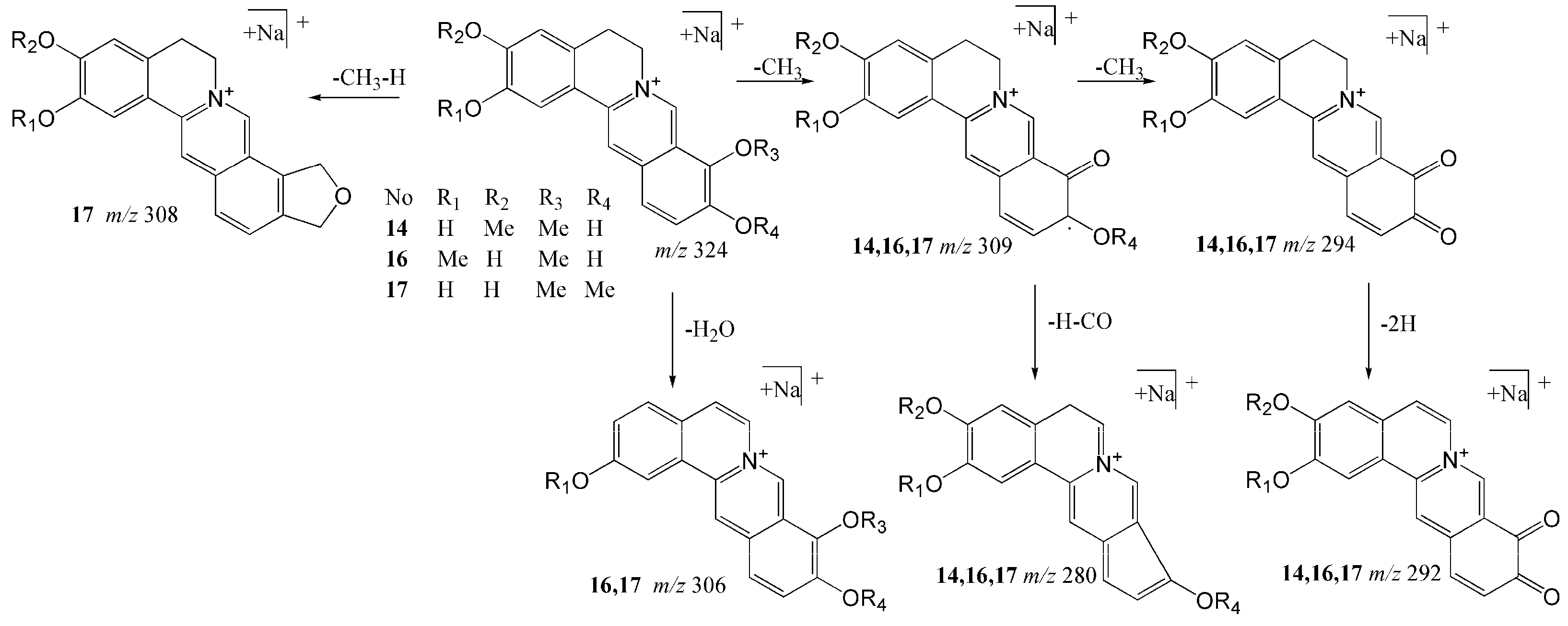
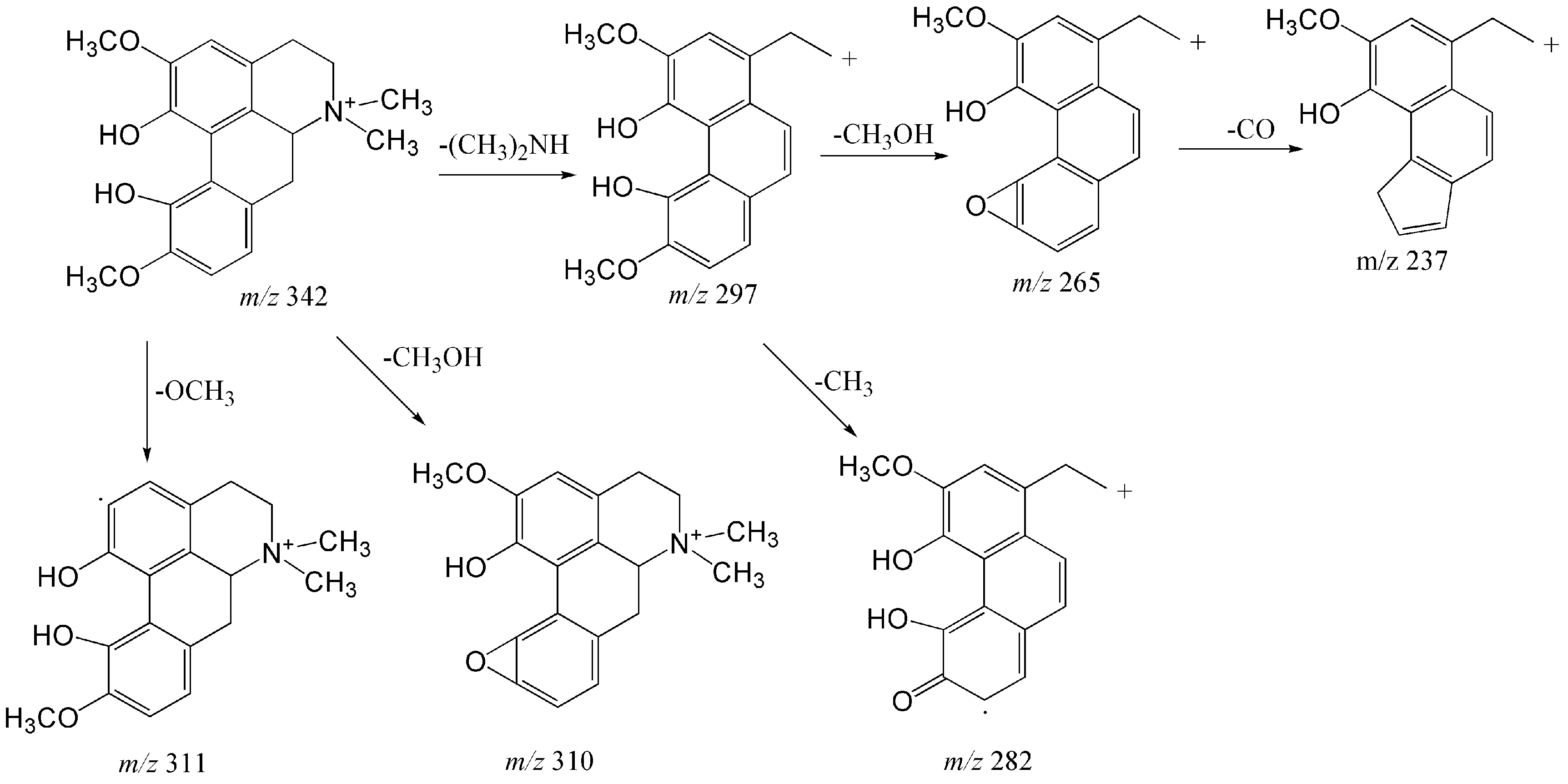
2.1.1. Structural Characterization and Identification of Alkaloids
2.1.2. Structural Characterization and Identification of Phenylpropanoids
2.1.3. Structural Characterization and Identification of Sesquiterpenoids
2.1.4. Structural Characterization and Identification of Other Compounds
2.2. Isolated Compounds Identification
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Sample and Standards Preparation
3.3. Extraction and Isolation
3.4. Instrumentation and Condition
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chinese Pharmacopeia Commission. Pharmacopeia of the People's Republic of China (Version 2015); China Medical Science Press: Beijing, China, 2015. [Google Scholar]
- Wu, F.R.; Zeng, C.Y.; Dai, W.B. Pharmacological Effects of Tinospora sinensis and Research Progress in its Clinical Application. China Licens. Pharm. 2014, 11, 37–40. [Google Scholar]
- Chi, S.S.; She, G.M.; Han, D.; Wang, W.H.; Liu, Z.; Liu, B. Genus Tinospora: Ethnopharmacology, Phytochemistry, and Pharmacology. Evid.-Based Complement. Altern. Med. 2016, 2, 1–32. [Google Scholar]
- Bai, W.T.; Xie, Y.H.; Wei, Y.F.; Huang, L.P. Phytochemical and Pharmacological Research Advances of Kuan Jin Teng, a Tibetan Medicinal Plants. Mod. Chin. Med. 2016, 18, 1666–1674. [Google Scholar]
- Maurya, R.; Gupta, P.; Chand, K.; Kumar, M.; Dixit, P.; Singh, N.; Dube, A. Constituents of Tinospora sinensis and their antileishmanial activity against Leishmania donovani. Nat. Prod. Res. 2009, 23, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Hegde, S.; Jayaraj, M. A Review of the Medicinal Properties, Phytochemical and Biological Active Compounds of Tinospora sinensis (Lour.) Merr. J. Biol. Act. Prod. Nat. 2016, 6, 84–94. [Google Scholar]
- Xu, L.L.; Guo, F.X.; Chi, S.S.; Wang, Z.J.; Jiang, Y.Y.; Liu, B.; Zhang, J.Y. Rapid screening and identification of diterpenoids in Tinospora sinensis based on high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry. Molecules 2017, 22, 912. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Gupta, P.; Maurya, U.R. A novel protein tyrosine phosphatase 1B inhibitor from Tinospora sinensis. Chron. Young Sci. 2012, 3, 199–203. [Google Scholar] [CrossRef]
- Punitha, D.; Udhayasankar, M.R.; Danya, U.; Arumugasamy, K.; Shalimol, A. Anti-inflammatory activity of characterized compound diosgenin isolated from Tinospora malabarica Miers in Ann. (Menispermaceae) in animal model. Int. J. Herb. Med. 2013, 1, 76–78. [Google Scholar]
- Patel, M.B.; Mishra, S.M. Magnoflorine from Tinospora cordifolia stem inhibits α-glucosidase and is antiglycemic in rats. J. Funct. Foods 2012, 4, 79–86. [Google Scholar] [CrossRef]
- Choi, J.; Shin, K.M.; Park, H.J.; Jung, H.J.; Kim, H.J.; Lee, Y.S.; Rew, J.H.; Lee, K.T. Anti-inflammatory and antinociceptive effects of sinapyl alcohol and its glucoside syringin. Planta Med. 2004, 70, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Xia, N. Syringin exhibits anticancer effects in HeLa human cervical cancer cells by inducing apoptosis, cell cycle arrest and inhibition of cell migration. Bangladesh J. Pharmacol. 2016, 11, 838–843. [Google Scholar] [CrossRef]
- Niu, H.S.; Liu, I.M.; Cheng, J.T.; Lin, C.L.; Hsu, F.L. Hypoglycemic effect of syringin from Eleutherococcus senticosus in streptozotocin-induced diabetic rats. Planta Med. 2008, 74, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Bala, M.; Kumar, N.; Singh, B.; Munshi, R.K.; Bhalerao, S. Immunomodulatory active compounds from Tinospora cordifolia. J. Ethnopharmacol. 2012, 141, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, X.J.; Xu, W.; Huang, J.; Zhu, D.Y.; Qiu, X.H. Rapid Characterization and Identification of Flavonoids in Radix Astragali by Ultra-High-Pressure Liquid Chromatography Coupled with Linear Ion Trap-Orbitrap Mass Spectrometry. J. Chromatogr. Sci. 2015, 53, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.J.; Zhang, J.Y.; Liu, Q.Q.; Wu, Z.S.; Liu, S.S.; Shi, H.; Ma, Q. Chemical profiling of fifty constituents in Qi-gui-yin granule (QGY) by on-line high-performance liquid chromatography coupled with ESI-LTQ-Orbitrap mass spectrometer. Res. J. Chem. Environ. 2013, 17, 16–27. [Google Scholar]
- Li, Y.; Liu, Y.; Liu, R.R.; Liu, S.Y.; Zhang, X.P.; Wang, Z.J.; Lu, J.Q. HPLC-LTQ-orbitrap MSn profiling method to comprehensively characterize multiple chemical constituents in xiao-er-qing-jie granules. Anal. Methods 2015, 7, 7511–7526. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Dong, X.; Liu, X.G.; Gao, W.; Li, P.; Yang, H. Rapid separation and identification of multiple constituents in Danhong Injection by ultra-high performance liquid chromatography coupled to electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Chin. J. Nat. Med. 2016, 14, 147–160. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Brown, M.; Baker, P.N.; Redman, C.W.G.; Kenny, L.C.; Kell, D.B. Metabolic profiling of serum using Ultra Performance Liquid Chromatography and the LTQ-Orbitrap mass spectrometry system. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 871, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.N.; Hao, H.P.; Wang, X.; Wu, X.L.; Wang, G.J.; Sang, G.W.; Liang, Y.; Xie, L.; Xia, C.H.; Yao, X.L. Diagnostic fragment-ion-based extension strategy for rapid screening and identification of serial components of homologous families contained in traditional Chinese medicine prescription using high-resolution LC-ESI-IT-TOF/MS: Shengmai injection as an example. J. Mass Spectrom. 2009, 44, 230–244. [Google Scholar] [PubMed]
- Wu, W.; Song, F.; Yan, C.; Liu, Z.; Liu, S. Structural analyses of protoberberine alkaloids in medicine herbs by using ESI-FT-ICR-MS and HPLC-ESI-MS(n). J. Pharm. Biomed. Anal. 2005, 37, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.Y.; Hashi, Y.; Chen, Z.L. Simultaneous detection of eight active components in Radix Tinosporae by ultra high performance liquid chromatography coupled with electrospray tandem mass spectrometry. J. Sep. Sci. 2016, 39, 2036–2042. [Google Scholar] [CrossRef] [PubMed]
- Qing, Z.X.; Cheng, P.; Zeng, J.G. Research progress on mass spectral fragmentation behaviour of alkaloids in Macleaya cordata. Chin. Tradit. Herb. Drugs 2013, 44, 2929–2939. [Google Scholar]
- Lu, X.R.; Qin, M.J.; Xu, D.R. Identification of Main Chemical Components of Tongguanwan Prescription by HPLC-ESI-MS/MS. Chin. J. Nat. Med. 2008, 6, 283–291. [Google Scholar] [CrossRef]
- Bajpai, V.; Singh, A.; Chandra, P.; Negi, M.P.S.; Kumar, N.; Kumar, B. Analysis of phytochemical variations in dioecious Tinospora cordifolia stems using HPLC/QTOF MS/MS and UPLC/QqQLIT-MS/MS. Phytochem. Anal. 2016, 27, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.P.; Chen, C.X.; Ni, W.; Xie, B.B.; Li, J.Z.; Liu, H.Y. A new dinorclerone diterpenoid glycoside from Tinospora sinensis. Nat. Prod. Res. 2010, 24, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhao, H.; Zou, L.S.; Liu, X.H.; Chai, C.; Wang, S.N.; Hua, Y.J. Chemical constituents of eucommiae Cortex by LC-triple TOF MS/MS. J. Chin. Mass Spectrom. Soc. 2017, 38, 146–156. [Google Scholar]
- Li, W.; Koike, K.; Liu, L.J.; Lin, L.B.; Fu, X.W.; Chen, Y.J.; Nikaido, T. New lignan glucosides from the stems of Tinospora sinensis. Chem. Pharm. Bull. 2004, 52, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, W.; Ma, F.H.; Li, Q.; Asada, Y.; Koike, K. Tinospinosides D, E, and tinospin E, further clerodane diterpenoids from Tinospora sagittata. Chem. Pharm. Bull. 2012, 60, 1324–1328. [Google Scholar] [CrossRef] [PubMed]
- Sabri, G.; Vishwakarma, R.A. Tinocordiside, a New Rearranged Cadinane Sesquiterpene Glycoside from Tinospora cordifolia. J. Nat. Prod. 1997, 60, 839–841. [Google Scholar]
- Maurya, R.; Handa, S.S. Tinocordifolin, a sesquiterpene from Tinospora cordifolia. Phytochemistry 1998, 49, 1343–1345. [Google Scholar] [CrossRef]
- Samita, F.; Ochieng, C.O.; Owuor, P.O.; Manguro, L.A.O. New ceramide from the aerial part of Tinospora oblongifolia with cytotoxic activities. Nat. Prod. Res. 2014, 28, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Xu, S.W.; Cai, L.; Zhou, B.; Chen, L.S. Research on Isolation of 2α-Hydroxy-oleanolic Acid and 2α-Hydroxy-ursolic Acid and Their EI-MS(+) Differences. Chem. Ind. For. Prod. 2010, 30, 91–94. [Google Scholar]
- Han, S.H.; Lee, H.H.; Lee, I.S.; Moon, Y.H.; Woo, E.R. A new phenolic amide from lycium chinense miller. Arch. Pharm. Res. 2002, 25, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.S.; Han, D.; Liu, B. Study on the determination method of total alkaloids in Tinospora sinensis caulis. Northwest Pharm. J. 2015, 30, 349–352. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
| NO. | tR/min | Identification | Experical Formula | Proposal Ions | Theoretical Mass m/z | Experimental Mass m/z | Mass Error (ppm) | MS2 Data (Measured) |
|---|---|---|---|---|---|---|---|---|
| 1 | 7.05 | Lotusine b | C19H24NO3 | [M]+ | 314.17507 | 314.17465 | −1.337 | 269 (100), 237 (8), 107 (12.85), 175 (15), 282 (4.7), 299(25) |
| 2 | 8.02 | 13-Hydroxy-2,3,9,10-tetramethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-α]-isoquinolinium *,b | C21H26NO5 | [M]+ | 372.18054 | 372.17999 | −1.503 | 192 (100), 177 (3), 176 (0.1), 208 (29), 165 (0.15), 356 (0.61), 354 (1.79) |
| 3 | 8.96 | Tembetarine | C20H26NO4 | [M]+ | 344.18563 | 344.18536 | −0.798 | 299 (100), 175 (44), 137 (17), 267 (12), 312 (11), 206 (2), 329 (3) |
| 4 | 9.41 | Magnoflorine | C20H24NO4 | [M]+ | 342.16998 | 342.16946 | −1.534 | 297 (100), 265 (23), 311 (16), 310 (1.5), 282 (2.6), 237 (1) |
| 5 | 11.02 | 2,11-dihydroxy-10-methoxy-6,6-dimethyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinolin-6-ium *,b | C22H24NO7 | [M − H + 2HCOOH]− | 414.15472 | 414.15561 | 2.128 | 354 (100), 368 (42), 353 (30), 386 (12), 369 (0.3) |
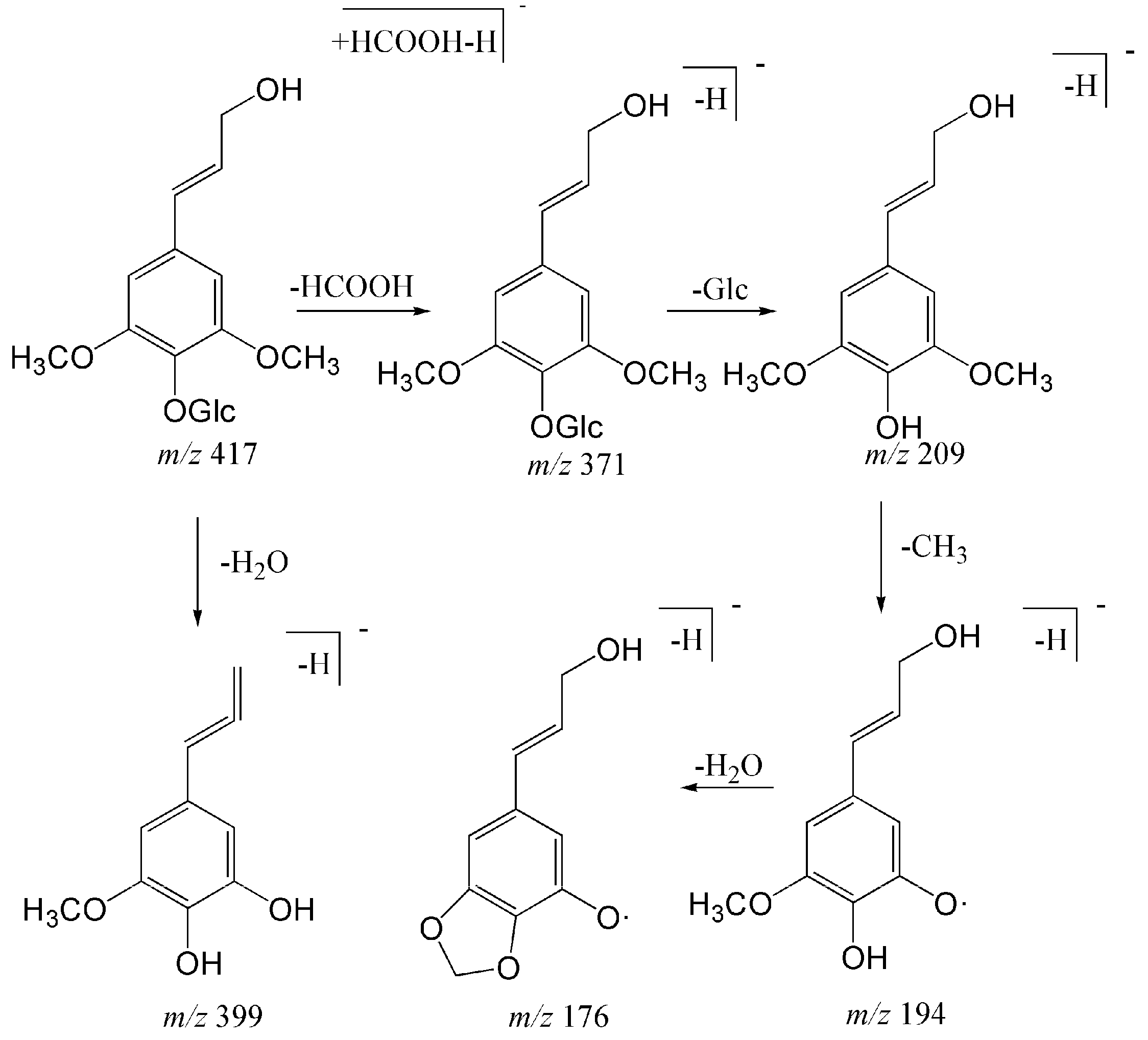
| 6 | 11.24 | Trans-syringin a | C18H25O11 | [M − H + HCOOH]− | 417.13913 | 417.13998 | 2.019 | 371 (3.3), 209 (100), 191 (10.62), 179 (12.32), 387 (4.76), 373 (10.54), 399 (13.63), 381 (7.27) |
| 7 | 11.40 | S-trans-N-methyltetra-hydrocolumbamine | C21H26NO4 | [M]+ | 356.18563 | 356.18527 | −1.024 | 192 (100), 177 (1.2), 190 (1), 325 (0.1) |
| 8 | 13.40 | Tinosinen | C22H32O13Na | [M + Na]+ | 527.17351 | 527.17303 | −0.914 | 317 (100), 496 (7), 395 (3.8), 233 (3) |
| 9 | 14.23 | Menisperine | C21H26NO4 | [M]+ | 356.18563 | 356.18555 | −0.238 | 311 (100), 279 (74), 251 (1.8), 325 (6.6), 296 (7.1) |
| 10 | 14.59 | Cyclanoline | C20H24NO4 | [M]+ | 342.16998 | 342.17026 | 0.805 | 192 (100), 190 (4), 177 (3.8), 311 (1.4) |
| 11 | 15.63 | Colletine b | C20H26NO3 | [M]+ | 328.19072 | 328.19031 | −1.250 | 283 (100), 251 (8), 175 (19), 143 (4), 144 (1), 296 (5.6), 313 (1.7) |
| 12 | 16.16 | Tanegoside | C27H35O14 | [M − H + HCOOH]− | 583.20213 | 583.20276 | 1.077 | 375 (100), 537 (17.89), 357 (0.32), 327 (18.28), 568 (0.59), 565 (0.34), 522 (0.25) |
| 13 | 18.02 | Tetrahydropamatine | C21H26NO4 | [M]+ | 356.18563 | 356.18588 | 0.688 | 192 (100), 177 (2.4), 190 (2), 165 (0.2), 340 (0.5) |
| 14 | 18.97 | Stepharanine | C19H18NO4 | [M]+ | 324.12303 | 324.12292 | −0.115 | 309 (100), 280 (2.2), 294 (0.34), 292 (0.16), 307 (3.2) |
| 15 | 19.39 | 2,3,9,10-Tetramethoxy-7-methyl-5,8,13,13a-tetrahydro-6H-isoquino[3,2-α]-isoquinolinium *,b | C22H28NO4 | [M]+ | 370.20128 | 370.20117 | −0.310 | 206 (100), 204 (2.2), 191 (1.5), 190 (1.47), 165 (0.24), 355 (0.38) |
| 16 | 19.95 | Dehydrodiscretamine | C19H18NO4 | [M]+ | 324.12303 | 324.12311 | 0.075 | 309 (100), 280 (1.2), 306 (0.17), 294 (0.08), 292 (0.05) |
| 17 | 20.36 | Demethyleneberberine | C19H18NO4 | [M]+ | 324.12303 | 324.12277 | −0.137 | 309 (100), 280 (11), 308 (9), 294 (0.8), 292 (0.56), 306 (0.22) |
| 18 | 21.15 | Pinoresinol-di-O-β-d-glucopyranoside b | C32H42O16Na | [M + Na]+ | 705.23650 | 705.23627 | −0.335 | 543 (100), 687 (1), 528 (0.58), 381 (0.16), 661 (0.16) |
| 19 | 22.62 | 13-Hydroxypalmatine | C21H22NO5 | [M]+ | 368.14924 | 368.14908 | −0.169 | 353 (100), 352 (99), 350 (20), 324 (21), 334 (0.6), 338 (1) |
| 20 | 24.27 | Icariside D1 | C19H28O10Na | [M + Na]+ | 439.15746 | 439.15729 | −0.406 | 307 (100), 421 (2.6), 275 (1.6), 403 (0.16), 407 (0.24) |
| 21 | 25.24 | 3-Hydroxy-2,9,11-trimethoxy-5,6-dihydro isoquino[3,2-α]-isoquinolinylium | C20H20NO4 | [M]+ | 338.13868 | 338.13861 | −0.075 | 323 (100), 295 (0.6), 308 (0.2), 294 (2), 320 (0.05) |
| 22 | 26.15 | 3(a,4-dihydroxy-3-methoxybenzyl)-4-(4-hydroxy-3-methoxybenzyl) tetrahydrofuran | C20H23O6 | [M − H]− | 359.14891 | 359.14923 | 0.315 | 341 (100), 344 (0.11), 329 (51), 311 (0.32), 205 (1.19) |
| 23 | 26.55 | Syringaresinol-di-O-β-d-glucoside b | C35H47O20 | [M − H + HCOOH]− | 787.26552 | 787.26581 | 0.368 | 579 (100), 417 (14), 769( 12), 741 (14), 723 (4) |
| 24 | 26.73 | Palmaturbine | C20H20NO4 | [M]+ | 338.13868 | 338.13837 | −0.315 | 323 (100), 295 (0.5), 308 (0.1), 294(2) |
| 25 | 27.41 | 2-{4-[4-(3,4-Dimethoxy-phenyl)-2,3-bis-hydroxymethylbutyl]-2-hydroxyphenoxy}-6-hydroxymethyl-tetrahydropyran-3,4,5-triol *,b | C26H36O11Na | [M + Na]+ | 547.21498 | 547.21509 | 0.107 | 532 (9.45), 385 (100), 367 (0.31), 349 (0.04), 514 (0.32), 517 (0.84), 529 (2.46), 531(1.5), 395(0.32) |
| 26 | 27.98 | Jatrorrhizine | C20H20NO4 | [M]+ | 338.13868 | 338.13834 | −0.345 | 323 (100), 322 (6), 295 (1), 308 (0.4), 294 (5), 320 (0.02) |
| 27 | 29.04 | Columbamine | C20H20NO4 | [M]+ | 338.13868 | 338.13843 | −0.255 | 323 (100), 322 (14), 308 (1.5), 295 (1), 294 (16) |
| 28 | 31.17 | 4-{4-[(3,4-Dimethoxy-phenyl)hydroxymethyl]-tetrahydrofuran-3-ylmethyl}-benzene-1,2-diol *,b | C20H23O6 | [M − H]− | 359.14891 | 359.14935 | 0.435 | 341 (100), 344 (0.28), 329 (48), 311 (1.41), 191 (0.22), 343 (0.06) |
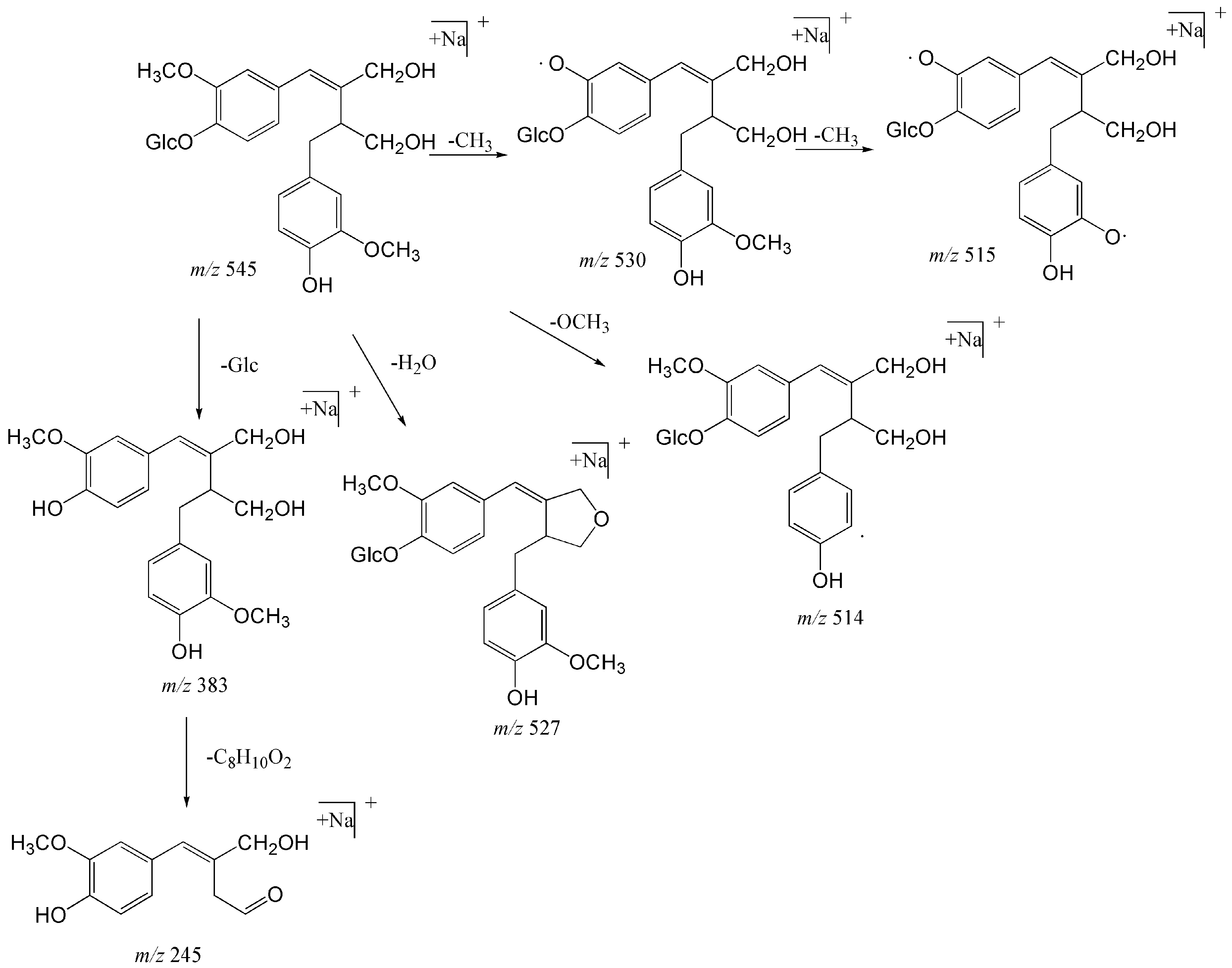
| 29 | 31.50 | Sagitiside A | C26H34O11Na | [M + Na]+ | 545.19933 | 545.19855 | −1.436 | 530 (100), 383 (34), 527 (14.88), 515 (11.30), 514 (3.30), 245 (1.32) |
| 30 | 31.97 | 8′-Epitanegool | C20H24O7Na | [M + Na]+ | 399.14142 | 399.14093 | −1.238 | 202 (100), 351 (37), 381 (33), 384 (22), 368 (8), 369 (9), 219 (7.5) |
| 31 | 33.68 | 4-[5-(4-Hydroxy-3-methoxyphenyl)-4-hydroxymethyl-tetrahydrofuran-3-ylmethyl]-6-methoxy-benzene-1,3-diol *,b | C20H24O7Na | [M + Na]+ | 399.14142 | 399.14133 | −0.236 | 202 (100), 351 (27), 381 (19.7), 384 (24), 369 (12), 368 (4), 219 (8.3) |
| 32 | 33.78 | Tinocordifolioside | C21H32O8Na | [M + Na]+ | 435.19893 | 435.19821 | −1.675 | 417 (20.57), 402 (0.13), 399 (0.78), 407 (8.28), 389 (1.93), 420 (0.53), 405 (0.77), 273 (100), 255 (2.15), 245 (0.15) |
| 33 | 33.97 | Berberine a | C20H18NO4 | [M]+ | 336.12303 | 336.12317 | 0.135 | 321 (100), 320 (25), 292 (21), 306 (1), 334 (0.66) |
| 34 | 35.15 | Palmatine | C21H22NO4 | [M]+ | 352.15433 | 352.1539 | −0.435 | 337 (69), 336 (100), 322 (0.5), 308 (20) |
| 35 | 37.33 | 6-{1-[3,4-Dihydroxy-6-hydroxymethyl-5-(3,4,5-trihydroxy-6-hydroxy-methyl-tetrahydropyran-2-yloxy)-tetrahydro-pyran-2-yloxy]-1-methylethyl}-2,12-dimethyltricyclo-[6.4.0.02,9]-dodec-11-en-10-one *,b | C27H42O12Na | [M + Na]+ | 581.25684 | 581.25641 | −0.753 | 365 (100), 347 (1.22), 563 (0.7), 551 (0.17), 533 (0.10), 419 (1.26), 401 (0.12), 257 (0.11) |
| 36 | 37.77 | 2-{4-[4-(4-Hydroxy-3-methoxyphenyl)-2,3-bis-hydroxymethylbutyl]-2-methoxyphenoxy}-6-hydroxymethyl-tetrahydro-pyran-3,4,5-triol *,b | C26H36O11Na | [M + Na]+ | 547.21498 | 547.21448 | −0.919 | 532 (100), 367 (42), 385 (35), 349 (8.6), 529 (13), 514 (5.8), 517 (2.5) |
| 37 | 38.60 | Pinoresinol-O-β-d-glucopyranoside | C26H31O11 | [M−H]− | 519.18608 | 519.1864 | 0.601 | 357 (100), 151 (0.21), 501 (0.04), 342 (0.08), 339 (0.02) |
| 38 | 38.69 | Tinocordifolin | C15H22O3Na | [M + Na]+ | 273.14611 | 273.14627 | 0.565 | 258 (11.24), 243 (10.32), 245 (27.57), 230 (4.6), 255 (100), 227 (27.93) |
| 39 | 39.57 | Syringaresinol-O-β-d-glucopyranoside | C28H36O13Na | [M + Na]+ | 603.20481 | 603.20416 | −1.081 | 441 (100), 573 (8.88), 588 (6.6), 585 (1.8), 426 (0.49), 423 (0.28) |
| 40 | 39.81 | 2-{4-[4-(3,5-Dimethoxy-phenyl)tetrahydro-furo[3,4-c]furan-1-yl]-2-methoxyphenoxy}-6-hydroxymethyl-tetrahydropyran-3,4,5-triol *,b | C28H35O13 | [M − H + HCOOH]− | 579.20721 | 579.20740 | 0.315 | 417 (100), 165 (0.08), 402 (0.08), 564 (0.04), 547 (0.02) |
| 41 | 40.47 | Lyoniresinol-2α-O-β-d-glucopyranoside | C28H38O13Na | [M + Na]+ | 605.22046 | 605.22021 | −0.252 | 443 (100), 425 (2), 413 (10), 395 (40.88), 412 (2.62), 590 (1.71), 576 (1.14), 574 (15.34), 587 (8.55) |
| 42 | 40.47 | 13-methylberberine b | C21H20NO4 | [M]+ | 350.13868 | 350.13852 | −0.47 | 335 (100), 334 (99), 306 (39), 332 (0.85), 320 (0.56) |
| 43 | 41.22 | 2-{4-[4-(3,4-Dimethoxy-phenyl)tetrahydro-furo[3,4-c]furan-1-yl]-2-hydroxyphenoxy}-6-hydroxymethyl-tetrahydropyran-3,4,5-triol *,b | C26H32O11Na | [M + Na]+ | 543.18368 | 543.18329 | −0.723 | 309 (100), 381 (38), 527 (25), 528 (24), 525 (17), 363 (7.8), 513 (8), 512 (6.49) |
| 44 | 41.23 | Tinosinenside | C26H40O11Na | [M + Na]+ | 551.24628 | 551.24579 | −0.895 | 335 (100), 503 (0.2), 521 (0.25), 533 (0.9), 419 (1.57), 387 (0.19), 257 (0.31), 239 (0.59), 203 (0.41) |
| 45 | 41.43 | 2-{4-[4-(3-Hydroxy-4,5-dimethoxyphenyl)-tetrahydrofuro[3,4-c]-furan-1-yl]-2,6-dimethoxy-phenoxy}-6-hydroxymethyl-tetrahydro-pyran-3,4,5-triol *,b | C28H36O13Na | [M + Na]+ | 603.20481 | 603.20441 | –0.667 | 441 (100), 573 (9.72), 588 (6.62), 585 (4.68), 426 (0.88), 423 (5.25), 587 (1) |
| 46 | 41.91 | Tinosposide A a | C27H35O11 | [M − H]− | 535.21738 | 535.21783 | 0.825 | 373 (100), 520 (1.14), 358 (4.6), 517 (1.05), 505 (0.39), 357 (0.33), 358 (3.92) |
| 47 | 41.97 | 3,9-Dihydroxy-megastigmane-3-O-β-d-glucopyranosyl(6→1)-β-d-xylpyranoside | C25H35O13 | [M − H + HCOOH]− | 543.20721 | 543.20740 | 0.336 | 528 (0.34), 497 (100), 525 (0.66), 507 (0.51), 411 (0.32) |
| 48 | 41.99 | N-trans-caffeoyltyramine a, b | C17H18NO4 | [M + H]+ | 300.12303 | 300.12320 | 0.551 | 163 (100), 138 (8.48), 135 (0.66), 121 (9.2) |
| 49 | 42.29 | 4-allyl-2-methoxyphenyl-6-O-β-d-glucopyranosyl-(6→1)-β-d-apiofuranoside | C21H30O11Na | [M + Na]+ | 481.16803 | 481.16748 | –1.149 | 349 (100), 317 (86), 440 (23), 291 (11), 466 (8) |
| 50 | 43.42 | Tinocordiside a | C21H32O7Na | [M + Na]+ | 419.20402 | 419.20309 | –2.229 | 401 (1.1), 391 (0.15), 375 (0.2), 257 (32.05), 239 (59.3), 203 (100), 185 (4.15) |
| 51 | 43.80 | Angelicoidenol-2-O-β-d-apiofuranosyl-(1→6)-β-d-glucopyranoside | C21H36O11Na | [M + Na]+ | 487.21498 | 487.21439 | –1.217 | 469 (5.76), 472 (0.37), 457 (1.55), 439 (1.50), 355 (100), 337 (0.12), 193 (0.12) |
| 52 | 49.24 | Pinoresinol monomethyl ether-O-β-d-gluco-pyranoside | C28H35O13 | [M − H + HCOOH]− | 579.20721 | 579.20703 | –0.324 | 563 (0.5), 371 (100), 533 (15.8), 417 (5.4), 561 (0.36), 399 (0.56) |
| 53 | 49.78 | N-p-Coumaroyltyramine b | C17H18NO3 | [M + H]+ | 284.12811 | 284.12802 | –0.352 | 147 (100), 138 (0.18), 121 (0.94), 119 (1.87) |
| 54 | 51.86 | N-trans-feruloyltyramine a | C18H20NO4 | [M + H]+ | 314.13868 | 314.13855 | –0.428 | 177 (100), 145 (7.8), 117 (0.89), 149 (0.14), 138 (0.02) |
| 55 | 70.84 | 3-Hydroxy-2,4,9,10-tetramethoxy-5,6-dihydro-isoquino[3,2-α]-isoquinolinylium *,b | C21H22NO5 | [M]+ | 368.14924 | 368.14905 | –0.199 | 352 (100), 353 (85), 337 (62), 322 (27), 321 (2.4), 350 (1.5), 338 (12), 324 (17) |
| 56 | 78.44 | 2-(4-Hydroxy-3-methoxy-benzyl)-3-(4-hydroxy-3-methoxy-benzylidene)-butane-1,4-diol *,b | C20H24O6Na | [M + Na]+ | 383.14650 | 383.14645 | –0.156 | 365 (100), 368 (43), 351 (12), 347 (10), 245 (1) |
| 57 | 80.17 | N-Formylannonain | C18H16NO3 | [M + H]+ | 294.11246 | 294.11221 | –0.884 | 249 (100), 219 (3.2), 264 (7), 266 (0.85), 236 (0.8) |
| 58 | 116.95 | Ursolic acid | C30H47O3 | [M − H]− | 455.35197 | 455.35257 | 1.314 | 437 (21.06), 419 (4.9), 411 (45.93), 397 (13.61), 410 (14.24), 407 (19.45) |
| 59 | 118.78 | Oleanic acid | C30H47O3 | [M − H]− | 455.35197 | 455.35291 | 2.060 | 437 (1.65), 419 (1.63), 411 (4.66), 397 (5.75), 410 (2.13), 407 (100) |
| 60 | 125.78 | β-Sitosterol glycoside | C36H61O8 | [M − H + HCOOH]− | 621.43609 | 621.43665 | 0.893 | 575 (100), 560 (21), 603 (26.74), 585 (15), 606 (8.33) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Q.-S.; Xu, L.-L.; Zhang, J.-Y.; Wang, Z.-J.; Jiang, Y.-Y.; Liu, B. Rapid Characterization and Identification of Non-Diterpenoid Constituents in Tinospora sinensis by HPLC-LTQ-Orbitrap MSn. Molecules 2018, 23, 274. https://doi.org/10.3390/molecules23020274
Jiao Q-S, Xu L-L, Zhang J-Y, Wang Z-J, Jiang Y-Y, Liu B. Rapid Characterization and Identification of Non-Diterpenoid Constituents in Tinospora sinensis by HPLC-LTQ-Orbitrap MSn. Molecules. 2018; 23(2):274. https://doi.org/10.3390/molecules23020274
Chicago/Turabian StyleJiao, Qi-Shu, Lu-Lu Xu, Jia-Yu Zhang, Zi-Jian Wang, Yan-Yan Jiang, and Bin Liu. 2018. "Rapid Characterization and Identification of Non-Diterpenoid Constituents in Tinospora sinensis by HPLC-LTQ-Orbitrap MSn" Molecules 23, no. 2: 274. https://doi.org/10.3390/molecules23020274






