MicroRNA and Transcriptomic Profiling Showed miRNA-Dependent Impairment of Systemic Regulation and Synthesis of Biomolecules in Rag2 KO Mice
Abstract
:1. Introduction
2. Material and Methods
2.1. Mice
2.2. Tissue Collection
2.3. RNA Isolation, Quality Check, and Microarray Analysis
2.4. Selection of Differentially Expressed miRNAs and Genes for Downstream Analysis
2.5. Prediction of Targets for Differentially Expressed miRNAs
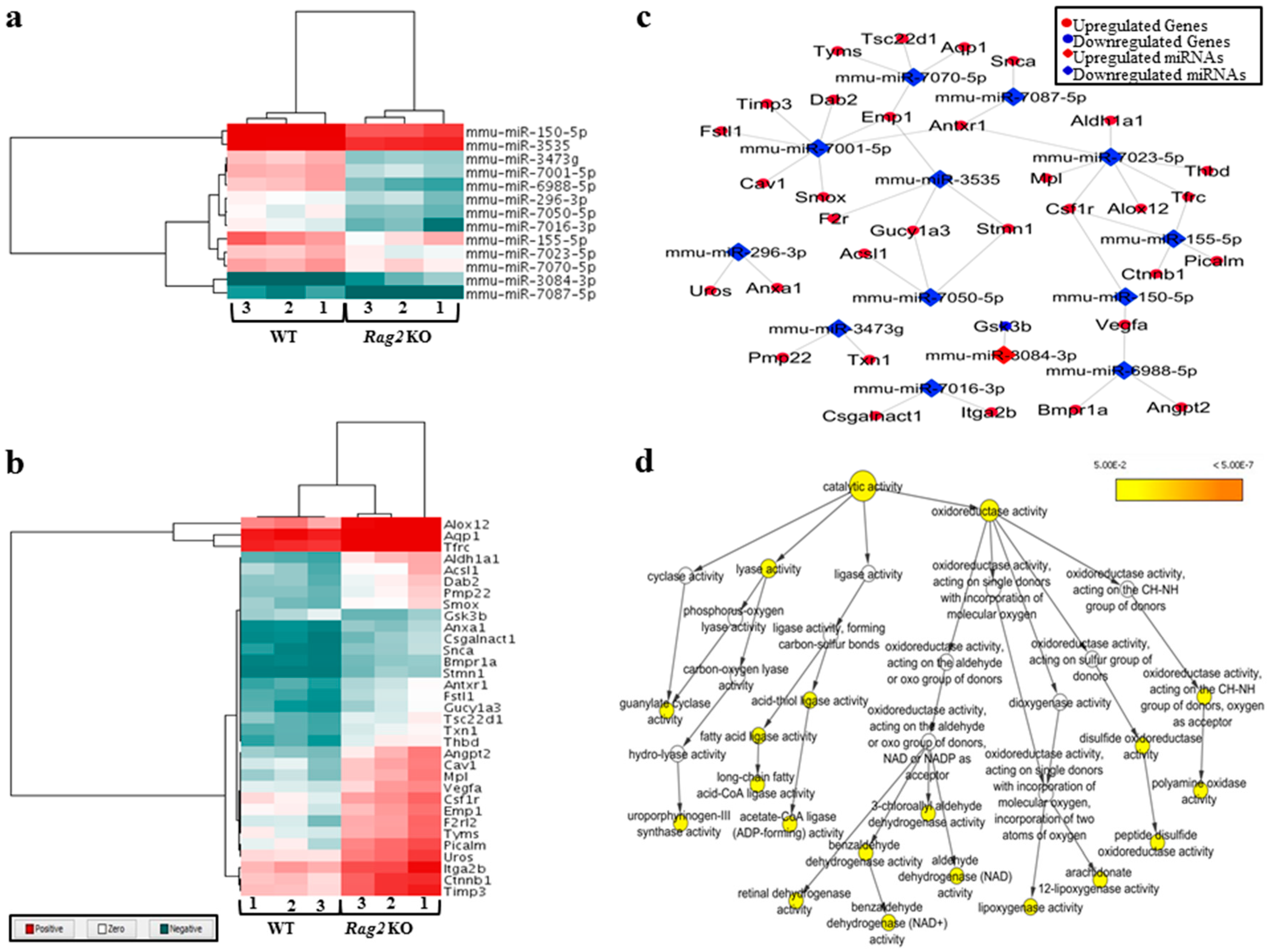
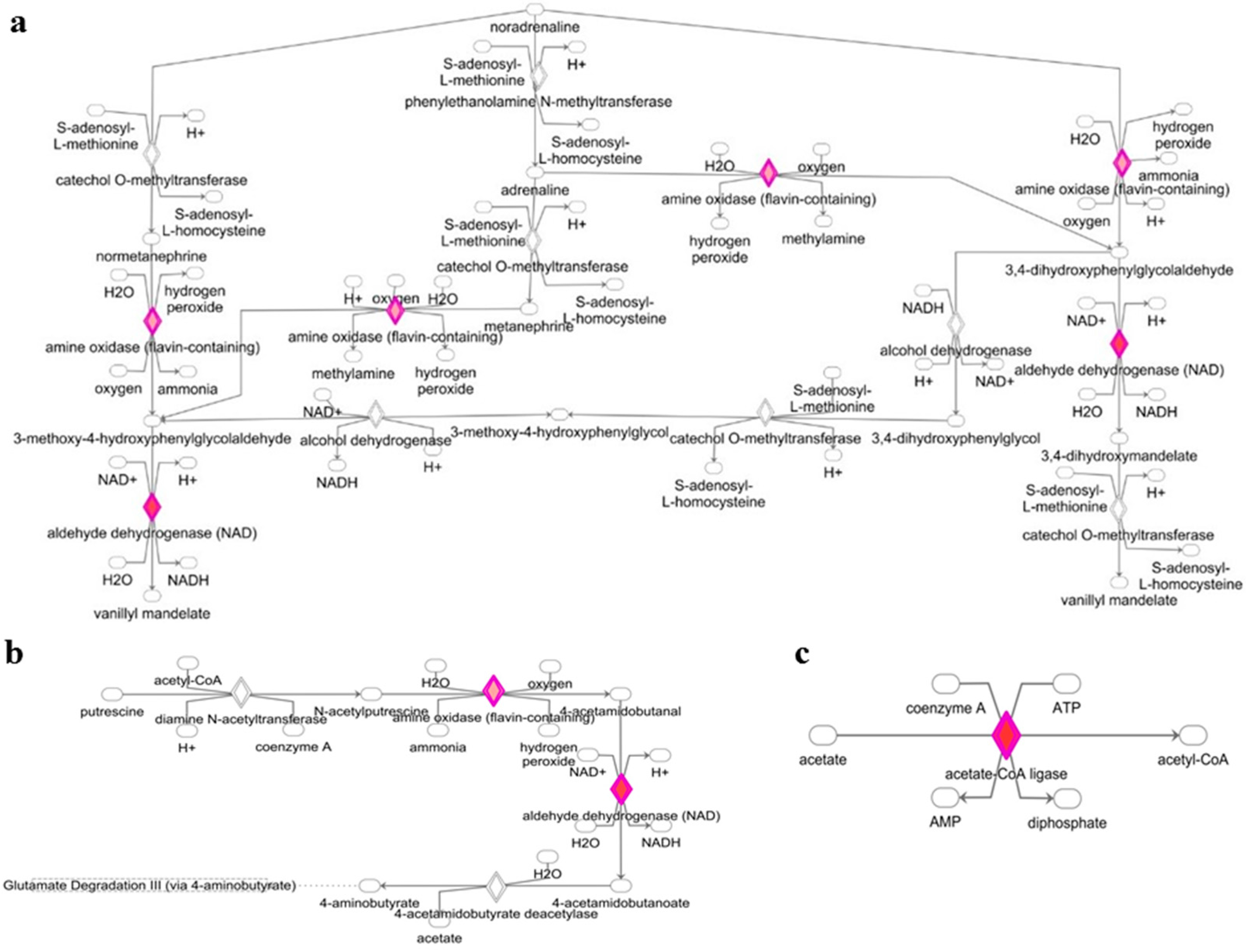
2.6. Generation of Heatmaps, miRNAs-Genes Interactinon Network and Catalytic Activity Networks
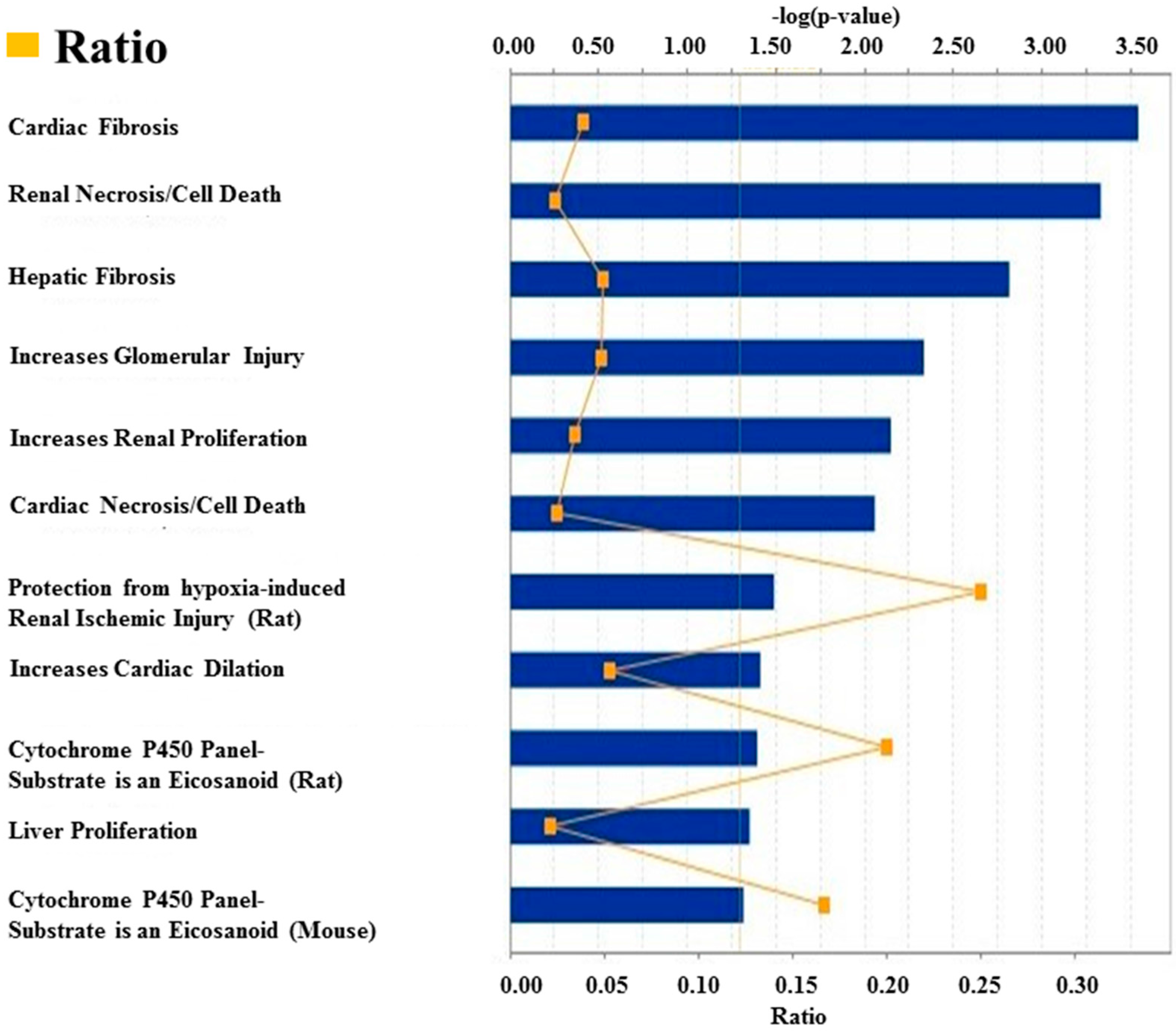
2.7. Ingenuity Pathway Analysis (IPA)-Based Canonical Pathways and Networks Detection
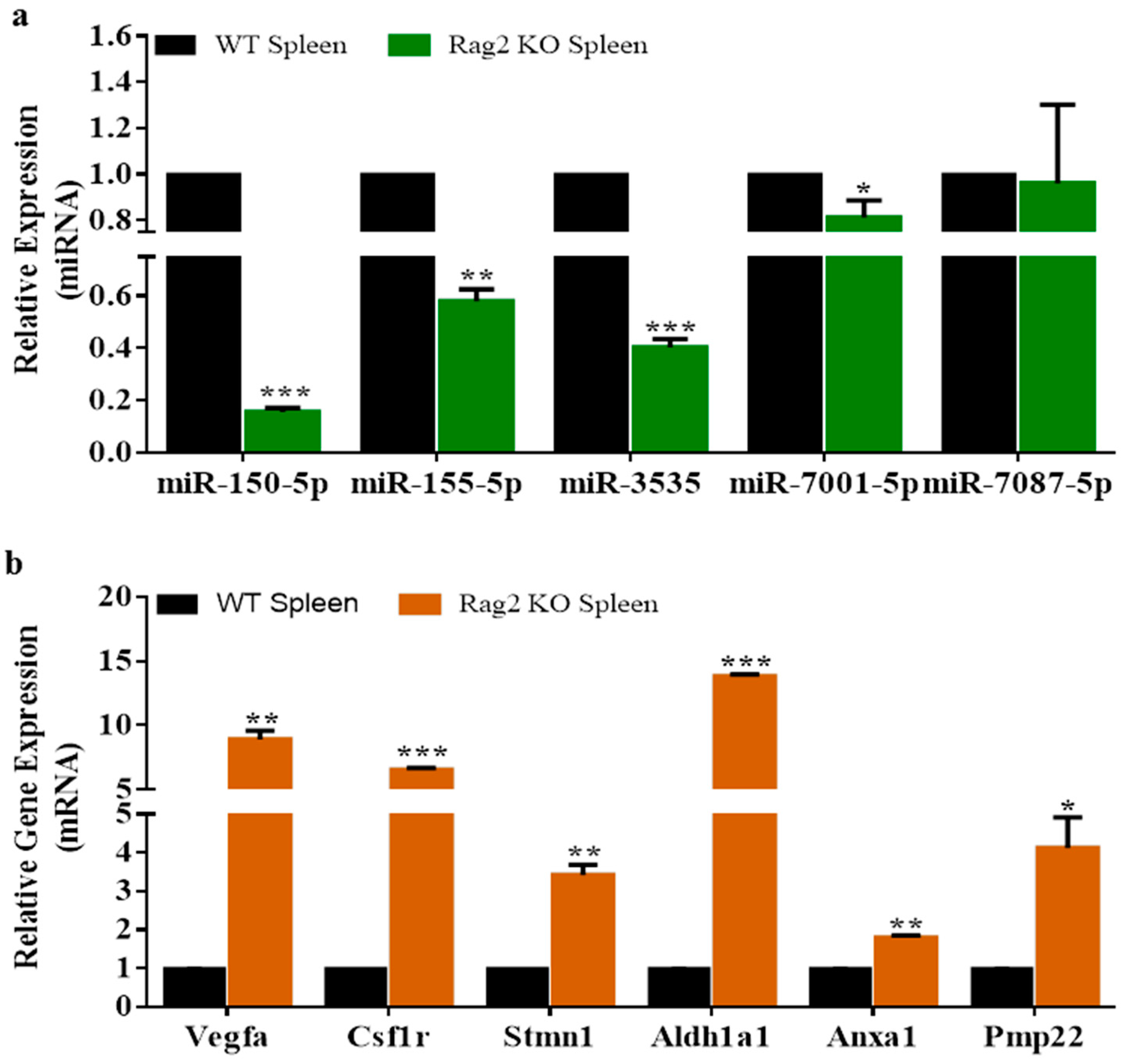
2.8. Validation of Deregulated miRNAs and Expression of Genes by qRT-PCR
2.9. Cut-Off Line and Statistical Analysis
3. Results
3.1. Deregulated miRNAs in Rag2 KO Mice Target Genes Involved in Systemic Regulation
3.2. Deregulated miRNAs in Rag2 KO Mice Target Genes Involved in Biomolecule Synthesis
3.3. Experimental Validation
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Reza, A.; Cho, S.K.; Choi, Y.J.; Hong, K.; Kim, J.H. Microarray profiling of mirna and mrna expression in Rag2 knockout and wild-type mouse spleens. Sci. Data 2018, 5, 170199. [Google Scholar] [CrossRef] [PubMed]
- Shinkai, Y.; Rathbun, G.; Lam, K.P.; Oltz, E.M.; Stewart, V.; Mendelsohn, M.; Charron, J.; Datta, M.; Young, F.; Stall, A.M.; et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 1992, 68, 855–867. [Google Scholar] [CrossRef]
- Choi, Y.-J.; Kim, E.; Reza, A.M.M.T.; Hong, K.; Song, H.; Park, C.; Cho, S.-K.; Lee, K.; Prather, R.S.; Kim, J.-H. Recombination activating gene-2(null) severe combined immunodeficient pigs and mice engraft human induced pluripotent stem cells differently. Oncotarget 2017, 8, 69398–69407. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.M.; Soroka, J.A.; Song, C.; Li, X.; Tonelli, L.H. Cd4+ t cells confer anxiolytic and antidepressant-like effects, but enhance fear memory processes in Rag2−/− mice. Stress 2016, 19, 303–311. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.O.; Hope, T.A.; Meck, W.H.; Kelsoe, G.; Williams, C.L. Impaired social recognition memory in recombination activating gene 1-deficient mice. Brain Res. 2011, 1383, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Craveiro, M.; Clerc, I.; Sitbon, M.; Taylor, N. Metabolic pathways as regulators of hiv infection. Curr. Opin. HIV AIDS 2013, 8, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Parvaneh, N.; Quartier, P.; Rostami, P.; Casanova, J.L.; de Lonlay, P. Inborn errors of metabolism underlying primary immunodeficiencies. J. Clin. Immun. 2014, 34, 753–771. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Semak, I.; Pisarchik, A.; Sweatman, T.; Szczesniewski, A.; Wortsman, J. Conversion of l-tryptophan to serotonin and melatonin in human melanoma cells. FEBS Lett. 2002, 511, 102–106. [Google Scholar] [CrossRef]
- Ikram, H.; Mushtaq, F.; Haleem, D.J. Dose-dependent effects of tryptophan on learning and memory. Pak. J. Pharm. Sci. 2014, 27, 1131–1135. [Google Scholar] [PubMed]
- Badawy, A.A. Tryptophan metabolism, disposition and utilization in pregnancy. Biosci. Rep. 2015, 35, e00261. [Google Scholar] [CrossRef] [PubMed]
- Radwanski, E.R.; Last, R.L. Tryptophan biosynthesis and metabolism: Biochemical and molecular genetics. Plant Cell 1995, 7, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Moehn, S.; Pencharz, P.B.; Ball, R.O. Lessons learned regarding symptoms of tryptophan deficiency and excess from animal requirement studies. J. Nutr. 2012, 142, 2231s–2235s. [Google Scholar] [CrossRef] [PubMed]
- Belizário, J.E. Immunodeficient mouse models: An overview. Open Immunol. J. 2009, 2, 79–85. [Google Scholar] [CrossRef]
- Reza, A.; Choi, Y.J.; Yuan, Y.G.; Das, J.; Yasuda, H.; Kim, J.H. Microrna-7641 is a regulator of ribosomal proteins and a promising targeting factor to improve the efficacy of cancer therapy. Sci. Rep. 2017, 7, 8365. [Google Scholar] [CrossRef] [PubMed]
- Fatima, F.; Nawaz, M. Vesiculated long non-coding rnas: Offshore packages deciphering trans-regulation between cells, cancer progression and resistance to therapies. Non-Coding RNA 2017, 3, 10. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. Clustermaker: A multi-algorithm clustering plugin for cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. Bingo: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Steimer, T. The biology of fear- and anxiety-related behaviors. Dialogues Clin. Neurosci. 2002, 4, 231–249. [Google Scholar] [PubMed]
- Mossio, M.; Montévil, M.; Longo, G. Theoretical principles for biology: Organization. Prog. Biophys. Mol. Biol. 2016, 122, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Loew, D. l-tryptophan. An essential amino acid for structural and functional metabolism. Fortschr. Med. 1997, 115, 40–42. [Google Scholar] [PubMed]
- Heine, W.; Radke, M.; Wutzke, K.D. The significance of tryptophan in human nutrition. Amino Acids 1995, 9, 91–205. [Google Scholar] [CrossRef] [PubMed]
- Fernstrom, J.D. Role of precursor availability in control of monoamine biosynthesis in brain. Physiol. Rev. 1983, 63, 484–546. [Google Scholar] [CrossRef] [PubMed]
- Schaechter, J.D.; Wurtman, R.J. Serotonin release varies with brain tryptophan levels. Brain Res. 1990, 532, 203–210. [Google Scholar] [CrossRef]
- Wurtman, R.J.; Anton-Tay, F. The mammalian pineal as a neuroendocrine transducer. Recent Prog. Horm. Res. 1969, 25, 493–522. [Google Scholar] [PubMed]
- Zhang, L.S.; Davies, S.S. Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 2016, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Tsuji, H.; Nakamura, S.; Ichiyama, A.; Nishizuka, Y.; Hayaishi, O. Studies on the biosynthesis of nicotinamide adenine dinucleotide. Ii. A role of picolinic carboxylase in the biosynthesis of nicotinamide adenine dinucleotide from tryptophan in mammals. J. Biol. Chem. 1965, 240, 1395–1401. [Google Scholar] [PubMed]
- Ledochowski, M.; Widner, B.; Murr, C.; Sperner-Unterweger, B.; Fuchs, D. Fructose malabsorption is associated with decreased plasma tryptophan. Scand. J. Gastroenterol. 2001, 36, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Ledochowski, M.; Sperner-Unterweger, B.; Widner, B.; Fuchs, D. Fructose malabsorption is associated with early signs of mental depression. Eur. J. Med. Res. 1998, 3, 295–298. [Google Scholar] [PubMed]
Sample Availability: Samples of the Rag2 KO mice are available from the authors. |


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reza, A.M.M.T.; Choi, Y.-J.; Kim, J.-H. MicroRNA and Transcriptomic Profiling Showed miRNA-Dependent Impairment of Systemic Regulation and Synthesis of Biomolecules in Rag2 KO Mice. Molecules 2018, 23, 527. https://doi.org/10.3390/molecules23030527
Reza AMMT, Choi Y-J, Kim J-H. MicroRNA and Transcriptomic Profiling Showed miRNA-Dependent Impairment of Systemic Regulation and Synthesis of Biomolecules in Rag2 KO Mice. Molecules. 2018; 23(3):527. https://doi.org/10.3390/molecules23030527
Chicago/Turabian StyleReza, Abu Musa Md Talimur, Yun-Jung Choi, and Jin-Hoi Kim. 2018. "MicroRNA and Transcriptomic Profiling Showed miRNA-Dependent Impairment of Systemic Regulation and Synthesis of Biomolecules in Rag2 KO Mice" Molecules 23, no. 3: 527. https://doi.org/10.3390/molecules23030527




