New Drug Candidate Targeting the 4A1 Orphan Nuclear Receptor for Medullary Thyroid Cancer Therapy
Abstract
:1. Introduction
2. Results
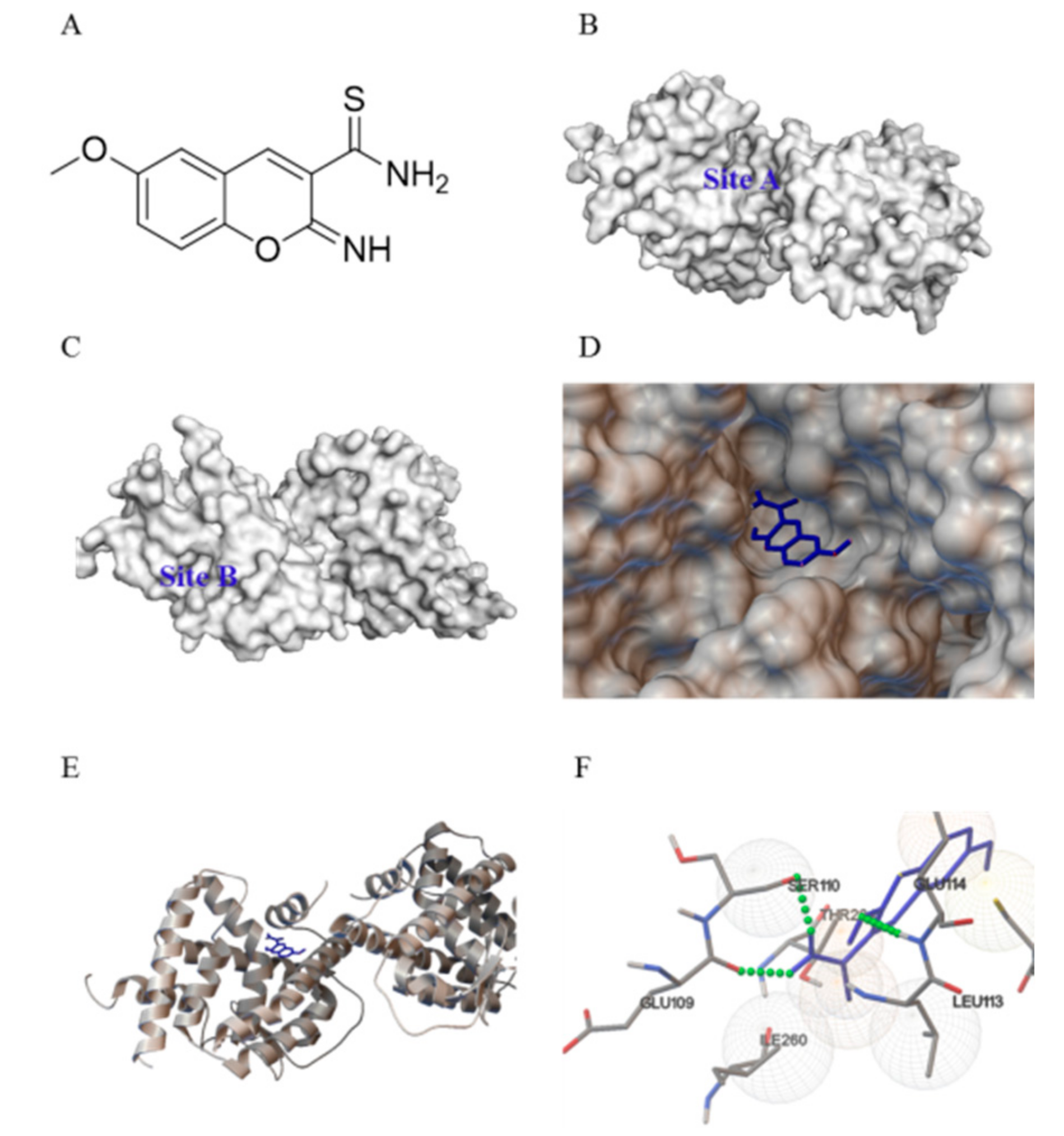
2.1. Computation-Based IMCA Binding and Interaction with NR4A1
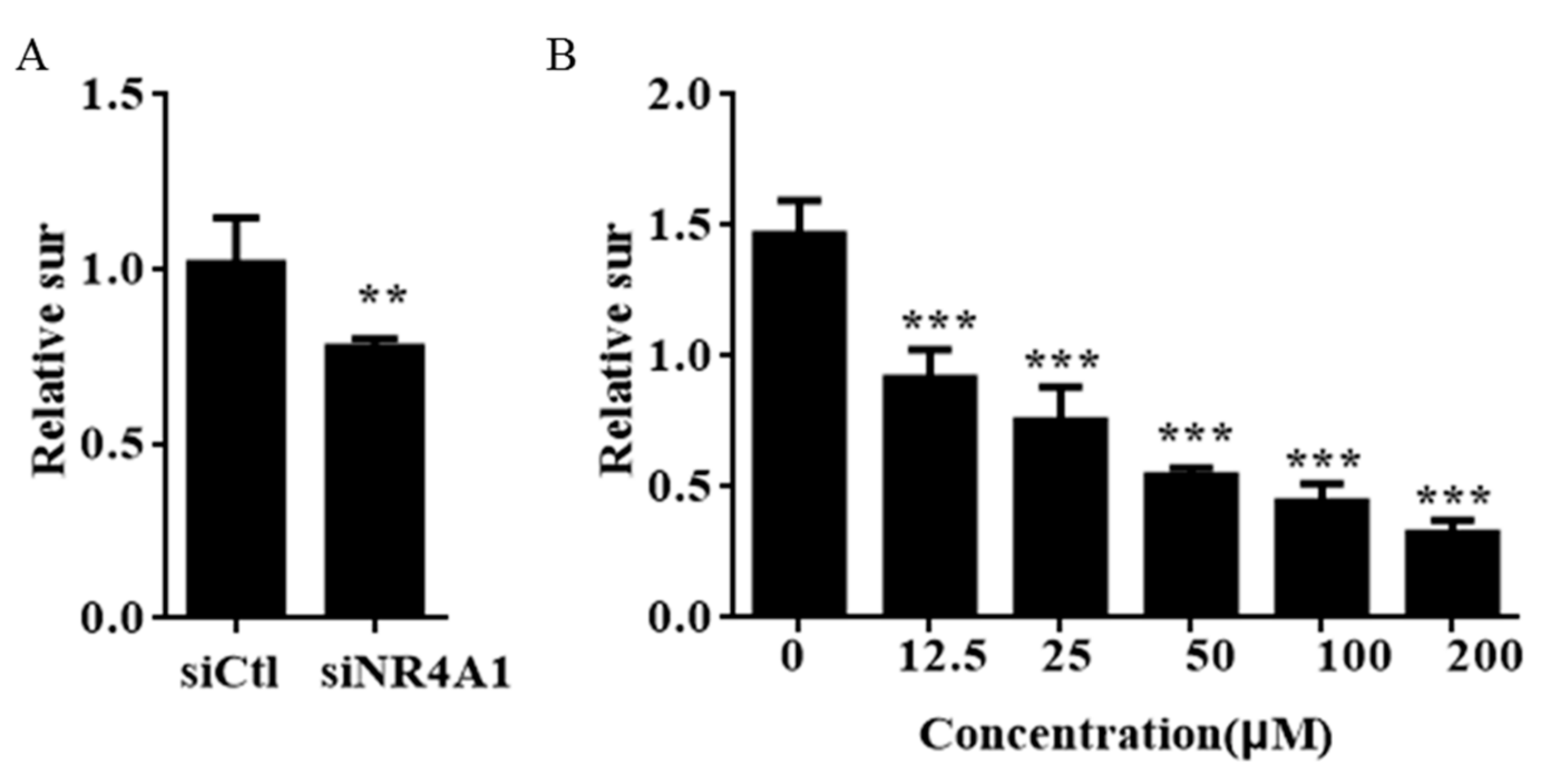
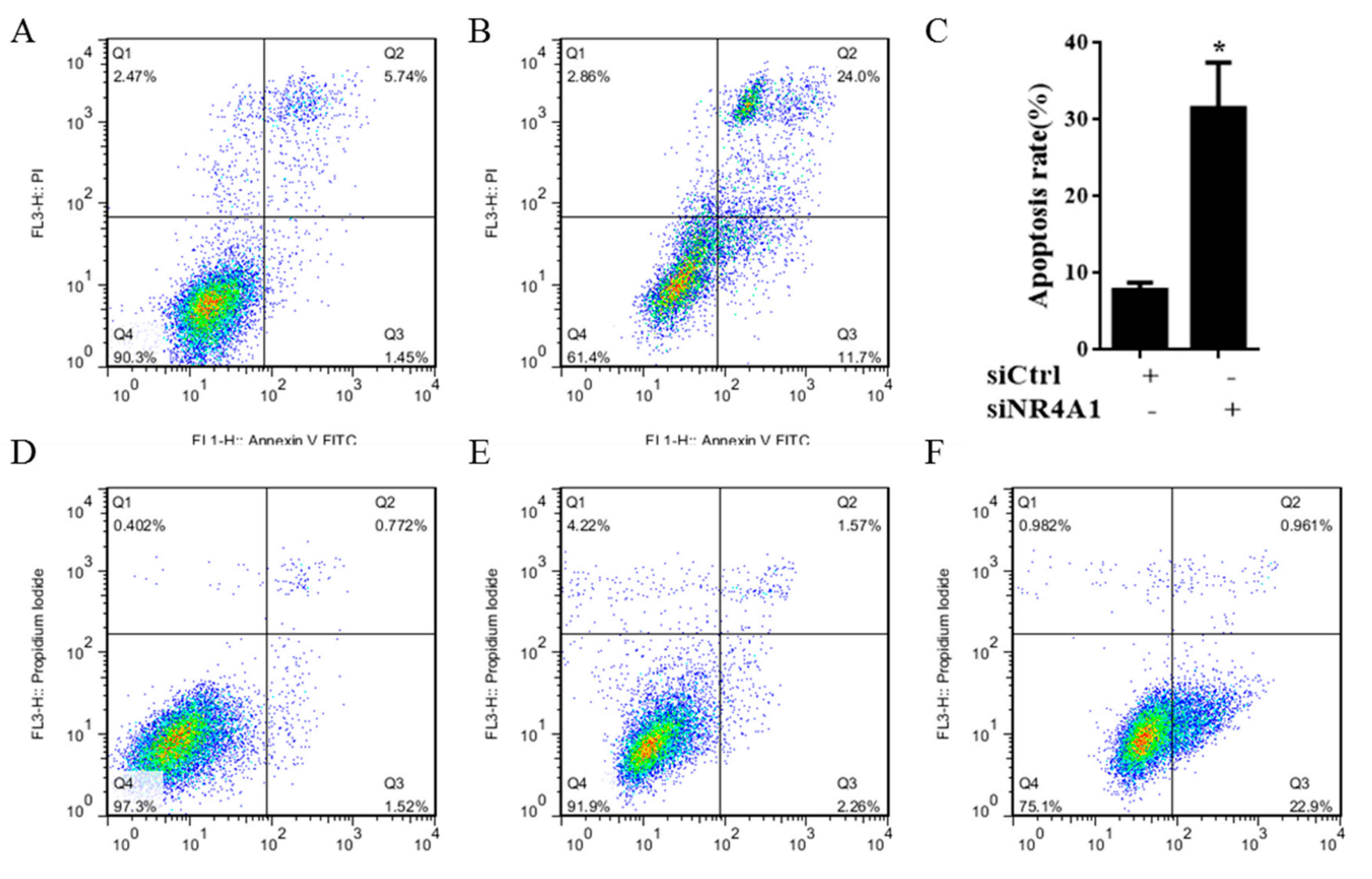
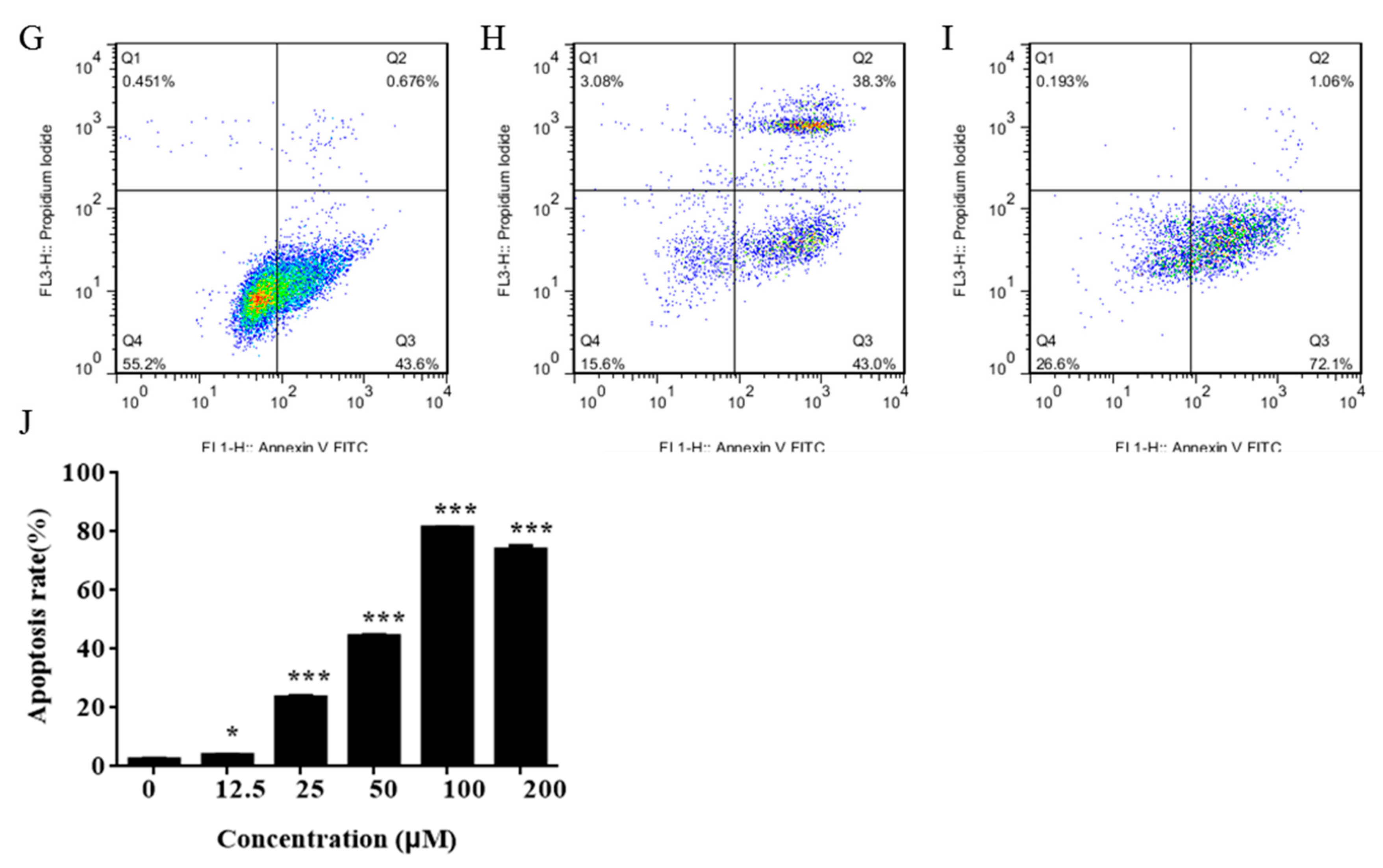
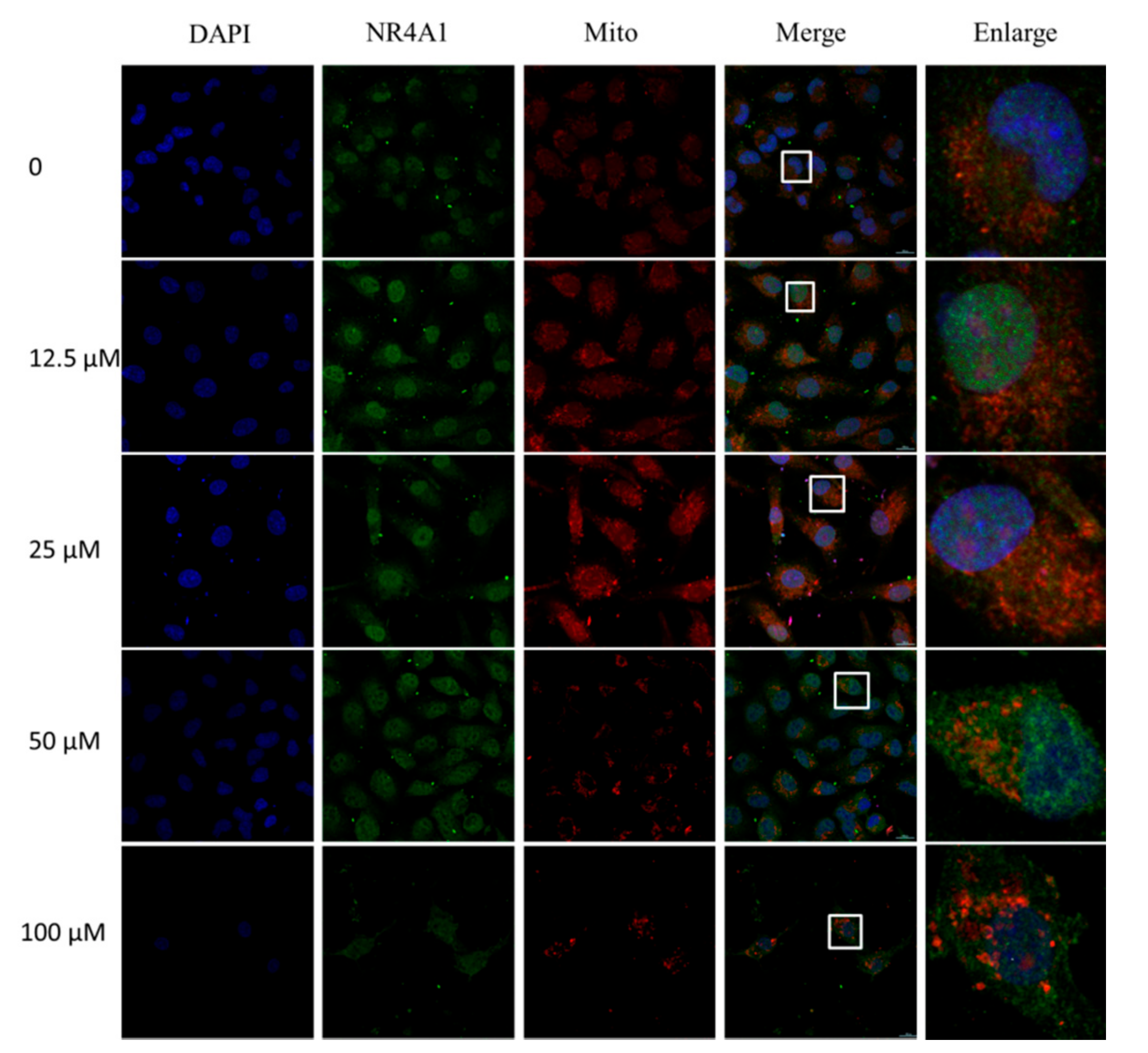
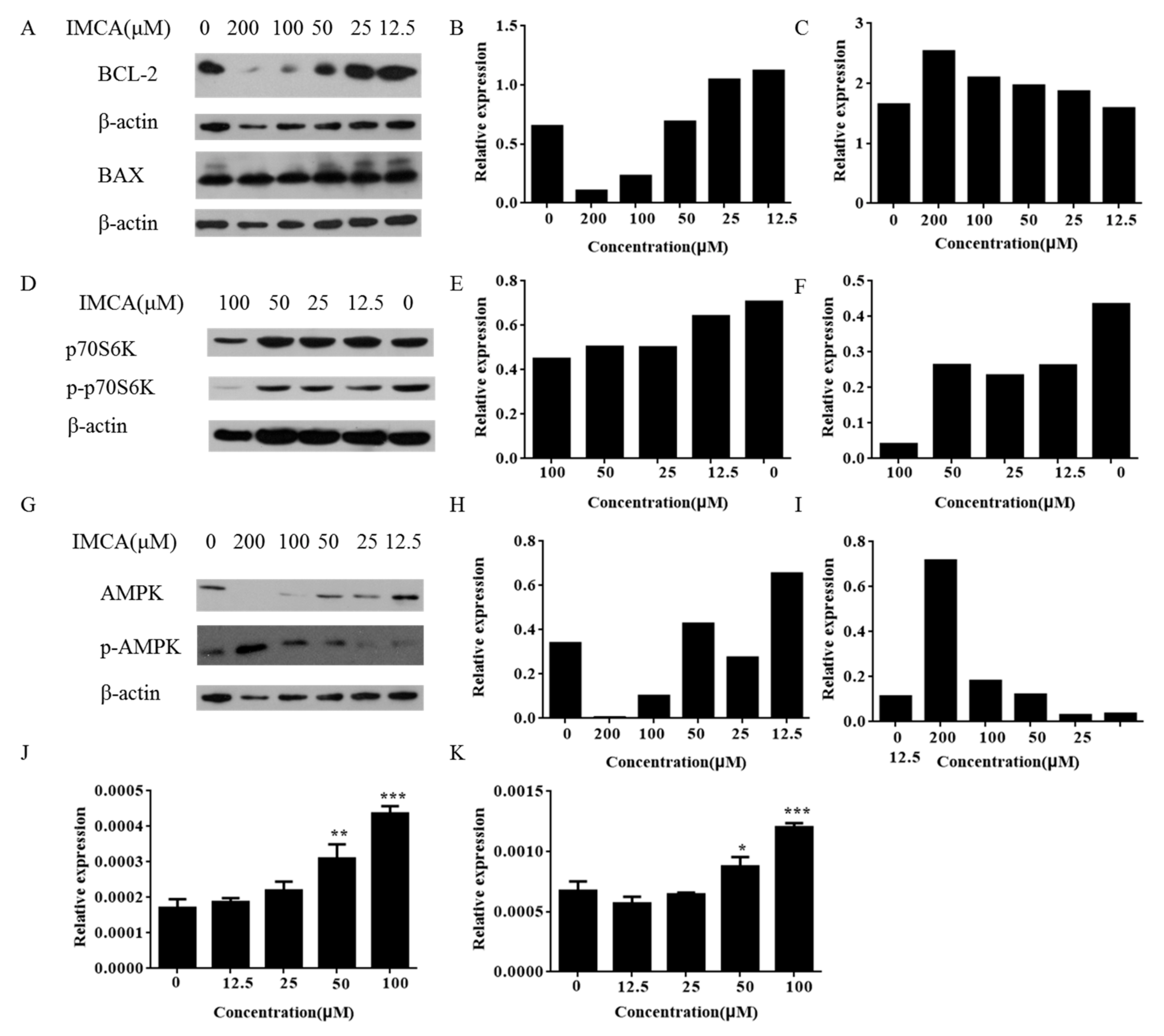
2.2. IMCA Inhibits Cell Proliferation and Induces Apoptosis in Thyroid Carcinoma Cells
2.3. IMCA Inhibits mTOR Signaling
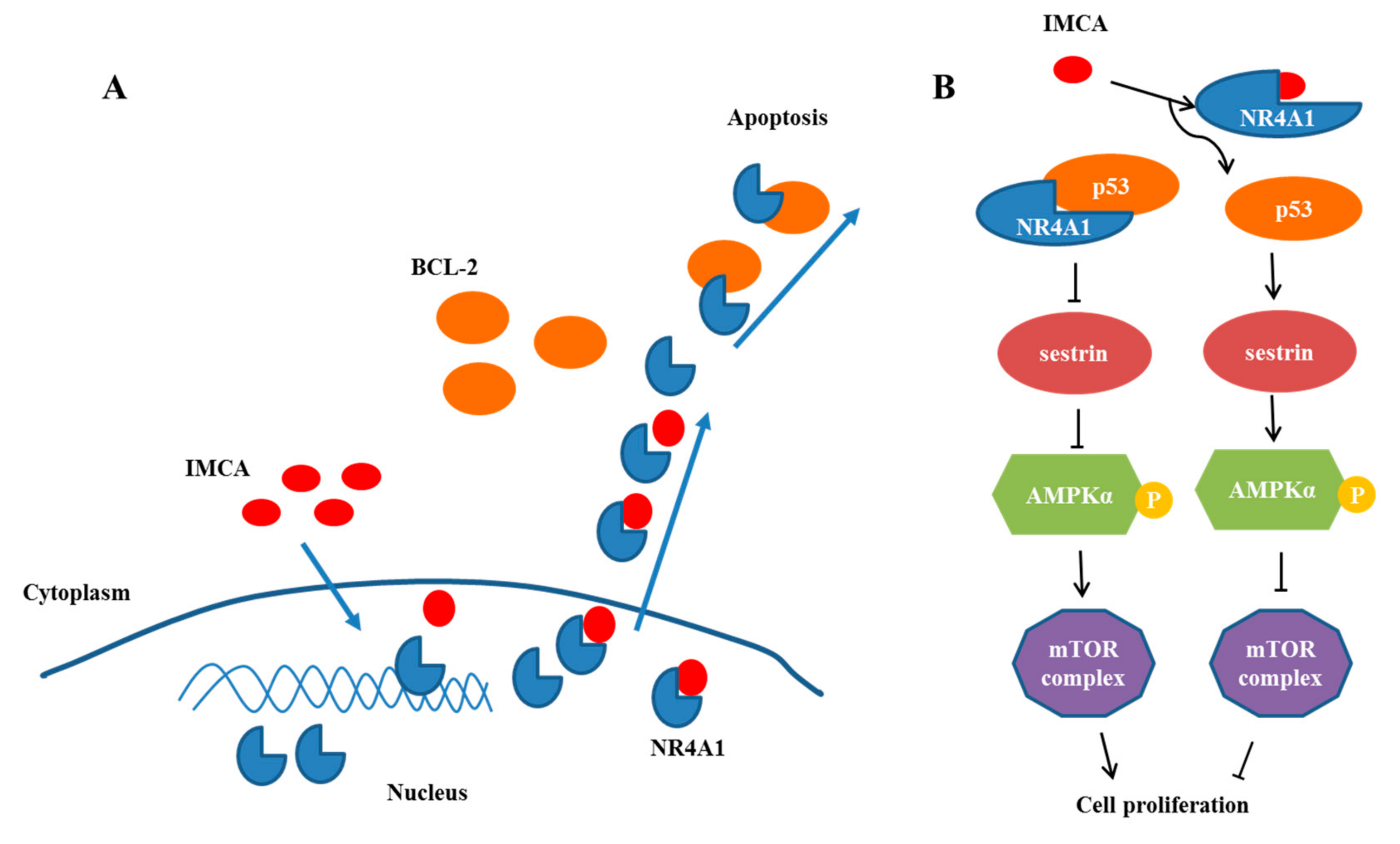
3. Discussion
4. Materials and Methods
4.1. Virtual Screening
4.2. Computation-Based Molecular Modeling
4.3. Chemicals and Reagents
4.4. Cell Lines and Cell Culture
4.5. siRNA Assay
4.6. MTT Assay
4.7. Annexin V and PI Staining
4.8. Immunofluorescence Assay
4.9. Real-Time PCR
4.10. Western Blotting
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nelkin, B. Recent advances in the biology and therapy of medullary thyroid carcinoma. F1000Research 2017, 6, 2184. [Google Scholar] [CrossRef] [PubMed]
- Thornton, K.; Kim, G.; Maher, V.E.; Chattopadhyay, S.; Tang, S.; Moon, Y.J.; Song, P.; Marathe, A.; Balakrishnan, S.; Zhu, H.; et al. Vandetanib for the treatment of symptomatic or progressive medullary thyroid cancer in patients with unresectable locally advanced or metastatic disease: U.S. Food and Drug Administration drug approval summary. Clin. Cancer Res. 2012, 18, 3722–3730. [Google Scholar] [CrossRef] [PubMed]
- Elisei, R.; Schlumberger, M.J.; Muller, S.P.; Schoffski, P.; Brose, M.S.; Shah, M.H.; Licitra, L.; Jarzab, B.; Medvedev, V.; Kreissl, M.C.; et al. Cabozantinib in progressive medullary thyroid cancer. J. Clin. Oncol. 2013, 31, 3639–3646. [Google Scholar] [CrossRef] [PubMed]
- Beard, J.A.; Tenga, A.; Chen, T. The interplay of NR4A receptors and the oncogene-tumor suppressor networks in cancer. Cell. Signal. 2015, 27, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, M.A.; Muscat, G.E. The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 2006, 4, e002. [Google Scholar] [CrossRef] [PubMed]
- Pearen, M.A.; Muscat, G.E. Minireview: Nuclear hormone receptor 4A signaling: Implications for metabolic disease. Mol. Endocrinol. 2010, 24, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.O.; Li, X.; Khan, S.; Safe, S. Targeting NR4A1 (TR3) in cancer cells and tumors. Expert Opin. Ther. Targets 2011, 15, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Moll, U.M.; Marchenko, N.; Zhang, X.K. p53 and Nur77/TR3-transcription factors that directly target mitochondria for cell death induction. Oncogene 2006, 25, 4725–4743. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.O.; Abdelrahim, M.; Yoon, K.; Chintharlapalli, S.; Papineni, S.; Kim, K.; Wang, H.; Safe, S. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res. 2010, 70, 6824–6836. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Kolluri, S.K.; Lin, F.; Liu, W.; Han, Y.H.; Cao, X.; Dawson, M.I.; Reed, J.C.; Zhang, X.K. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell 2004, 116, 527–540. [Google Scholar] [CrossRef]
- Kolluri, S.K.; Zhu, X.; Zhou, X.; Lin, B.; Chen, Y.; Sun, K.; Tian, X.; Town, J.; Cao, X.; Lin, F.; et al. A short Nur77-derived peptide converts Bcl-2 from a protector to a killer. Cancer Cell 2008, 14, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kim, S.G.; Blenis, J. Rapamycin: One drug, many effects. Cell Metab. 2014, 19, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Liu, P.; Wei, W. mTOR signaling in tumorigenesis. Biochim. Biophys. Acta 2014, 1846, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.O.; Andey, T.; Jin, U.H.; Kim, K.; Singh, M.; Safe, S. The nuclear receptor TR3 regulates mTORC1 signaling in lung cancer cells expressing wild-type p53. Oncogene 2012, 31, 3265–3276. [Google Scholar] [CrossRef] [PubMed]
- Chantranupong, L.; Wolfson, R.L.; Orozco, J.M.; Saxton, R.A.; Scaria, S.M.; Bar-Peled, L.; Spooner, E.; Isasa, M.; Gygi, S.P.; Sabatini, D.M. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Yin, N.; Li, M.O. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell 2014, 159, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Benoit, G.; Liu, J.; Prasad, S.; Aarnisalo, P.; Liu, X.; Xu, H.; Walker, N.P.; Perlmann, T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 2003, 423, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Du, X.; Chen, H.; Liu, J.; Zhao, B.; Huang, D.; Li, G.; Xu, Q.; Zhang, M.; Weimer, B.C.; et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat. Chem. Biol. 2008, 4, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Zeng, H.N.; Zhang, L.R.; Zhan, Y.Y.; Chen, Y.; Wang, Y.; Wang, J.; Xiang, S.H.; Liu, W.J.; Wang, W.J.; et al. A unique pharmacophore for activation of the nuclear orphan receptor Nur77 in vivo and in vitro. Cancer Res. 2010, 70, 3628–3637. [Google Scholar] [CrossRef] [PubMed]
- Chintharlapalli, S.; Burghardt, R.; Papineni, S.; Ramaiah, S.; Yoon, K.; Safe, S. Activation of Nur77 by selected 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J. Biol. Chem. 2005, 280, 24903–24914. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.D.; Yoon, K.; Chintharlapalli, S.; Abdelrahim, M.; Lei, P.; Hamilton, S.; Khan, S.; Ramaiah, S.K.; Safe, S. Nur77 agonists induce proapoptotic genes and responses in colon cancer cells through nuclear receptor-dependent and nuclear receptor-independent pathways. Cancer Res. 2007, 67, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, E.; Lee, S.O.; Doddapaneni, R.; Singh, M.; Safe, S. Nuclear receptor 4A1 as a drug target for breast cancer chemotherapy. Endocr.-Relat. Cancer 2015, 22, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.Y.; Chen, Y.; Zhang, Q.; Zhuang, J.J.; Tian, M.; Chen, H.Z.; Zhang, L.R.; Zhang, H.K.; He, J.P.; Wang, W.J.; et al. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat. Chem. Biol. 2012, 8, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.Y.; Yue, P.; Chandraratna, R.A.; Tesfaigzi, Y.; Hong, W.K.; Lotan, R. Dual mechanisms of action of the retinoid CD437: Nuclear retinoic acid receptor-mediated suppression of squamous differentiation and receptor-independent induction of apoptosis in UMSCC22B human head and neck squamous cell carcinoma cells. Mol. Pharmacol. 2000, 58, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhao, W.; Xie, G.; Huang, M.; Hu, M.; Jiang, X.; Zeng, D.; Liu, J.; Zhou, H.; Chen, H.; et al. Induction of Nur77-dependent apoptotic pathway by a coumarin derivative through activation of JNK and p38 MAPK. Carcinogenesis 2014, 35, 2660–2669. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Jin, U.H.; Morpurgo, B.; Abudayyeh, A.; Singh, M.; Tjalkens, R.B. Nuclear receptor 4A (NR4A) family - orphans no more. J. Steroid Biochem. Mol. Biol. 2016, 157, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Ferlini, C.; Cicchillitti, L.; Raspaglio, G.; Bartollino, S.; Cimitan, S.; Bertucci, C.; Mozzetti, S.; Gallo, D.; Persico, M.; Fattorusso, C.; et al. Paclitaxel directly binds to Bcl-2 and functionally mimics activity of Nur77. Cancer Res. 2009, 69, 6906–6914. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lin, Y.; Li, W.; Sun, Z.; Gao, W.; Zhang, H.; Xie, L.; Jiang, F.; Qin, B.; Yan, T.; et al. Regulation of Nur77 expression by beta-catenin and its mitogenic effect in colon cancer cells. FASEB J. 2011, 25, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.D.; Lee, S.O.; Chintharlapalli, S.; Abdelrahim, M.; Khan, S.; Yoon, K.; Kamat, A.M.; Safe, S. Activation of nerve growth factor-induced B alpha by methylene-substituted diindolylmethanes in bladder cancer cells induces apoptosis and inhibits tumor growth. Mol. Pharmacol. 2010, 77, 396–404. [Google Scholar] [PubMed]
- Lee, S.O.; Jin, U.H.; Kang, J.H.; Kim, S.B.; Guthrie, A.S.; Sreevalsan, S.; Lee, J.S.; Safe, S. The orphan nuclear receptor NR4A1 (Nur77) regulates oxidative and endoplasmic reticulum stress in pancreatic cancer cells. Mol. Cancer Res. 2014, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Wang, Y.; Chen, H.Z.; Xing, Y.Z.; Li, F.W.; Zhang, Q.; Zhou, B.; Zhang, H.K.; Zhang, J.; Bian, X.L.; et al. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nat. Chem. Biol. 2014, 10, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.O.; Li, X.; Hedrick, E.; Jin, U.H.; Tjalkens, R.B.; Backos, D.S.; Li, L.; Zhang, Y.; Wu, Q.; Safe, S. Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Mol. Endocrinol. 2014, 28, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Liu, A.Y.; Roland, J.; Adebayo, O.B.; Fletcher, S.A.; Slaughter, J.C.; Saskowski, J.; Crispens, M.A.; Jones, H.W.; James, S.; et al. TR3 modulates platinum resistance in ovarian cancer. Cancer Res. 2013, 73, 4758–4769. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.X.; Chen, H.Z.; Lei, N.Z.; Li, G.D.; Zhao, W.X.; Zhan, Y.Y.; Liu, B.; Lin, S.C.; Wu, Q. p53 mediates the negative regulation of MDM2 by orphan receptor TR3. EMBO J. 2006, 25, 5703–5715. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Campone, M.; Piccart, M.; Burris, H.A.; Rugo, H.S.; Sahmoud, T.; Noguchi, S.; Gnant, M.; Pritchard, K.I.; Lebrun, F.; et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012, 366, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Ciruelos Gil, E.M. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treat. Rev. 2014, 40, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.; Tan, Y.; Qu, K.; He, X.; Zhang, H.; Wang, Z. Diallyl disulphide inhibits apolipoprotein(a) expression in HepG2 cells through the MEK1-ERK1/2-ELK-1 pathway. Lipids Health Dis. 2017, 16, 223. [Google Scholar] [CrossRef] [PubMed]
- Tiedje, V.; Stuschke, M.; Weber, F.; Dralle, H.; Moss, L.; Fuhrer, D. Anaplastic thyroid carcinoma: Review of treatment protocols. Endocr.-Relat. Cancer 2018, 25, R153–R161. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Wang, Y.; Zhang, X.; Hu, Z.; Yuan, M.; Yang, K. Autographa californica Nucleopolyhedrovirus Ac76: A dimeric type II integral membrane protein that contains an inner nuclear membrane-sorting motif. J. Virol. 2014, 88, 1090–1103. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Liu, W.; Wang, Q.; Li, Q.; Wang, H.; Wang, J.; Teng, T.; Chen, M.; Ji, A.; Li, Y. New Drug Candidate Targeting the 4A1 Orphan Nuclear Receptor for Medullary Thyroid Cancer Therapy. Molecules 2018, 23, 565. https://doi.org/10.3390/molecules23030565
Zhang L, Liu W, Wang Q, Li Q, Wang H, Wang J, Teng T, Chen M, Ji A, Li Y. New Drug Candidate Targeting the 4A1 Orphan Nuclear Receptor for Medullary Thyroid Cancer Therapy. Molecules. 2018; 23(3):565. https://doi.org/10.3390/molecules23030565
Chicago/Turabian StyleZhang, Lei, Wen Liu, Qun Wang, Qinpei Li, Huijuan Wang, Jun Wang, Tieshan Teng, Mingliang Chen, Ailing Ji, and Yanzhang Li. 2018. "New Drug Candidate Targeting the 4A1 Orphan Nuclear Receptor for Medullary Thyroid Cancer Therapy" Molecules 23, no. 3: 565. https://doi.org/10.3390/molecules23030565





