Identification and Characterizations of Novel, Selective Histone Methyltransferase SET7 Inhibitors by Scaffold Hopping- and 2D-Molecular Fingerprint-Based Similarity Search
Abstract
:1. Introduction
2. Results and Discussions
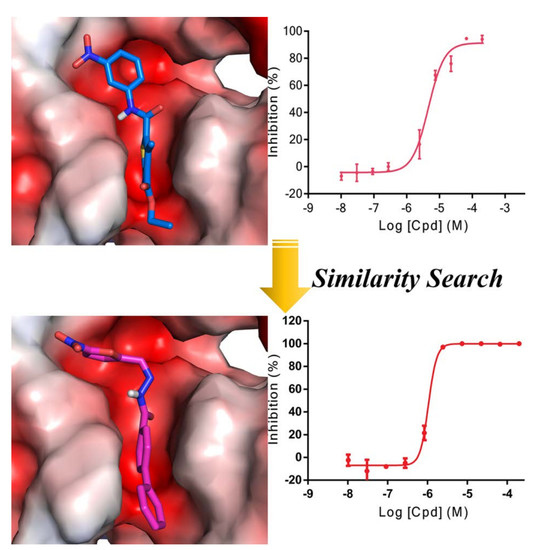
2.1. Scaffold Hopping- and Similarity-Based Virtual Screening
2.2. AlphaLISA-Based Biological Tests
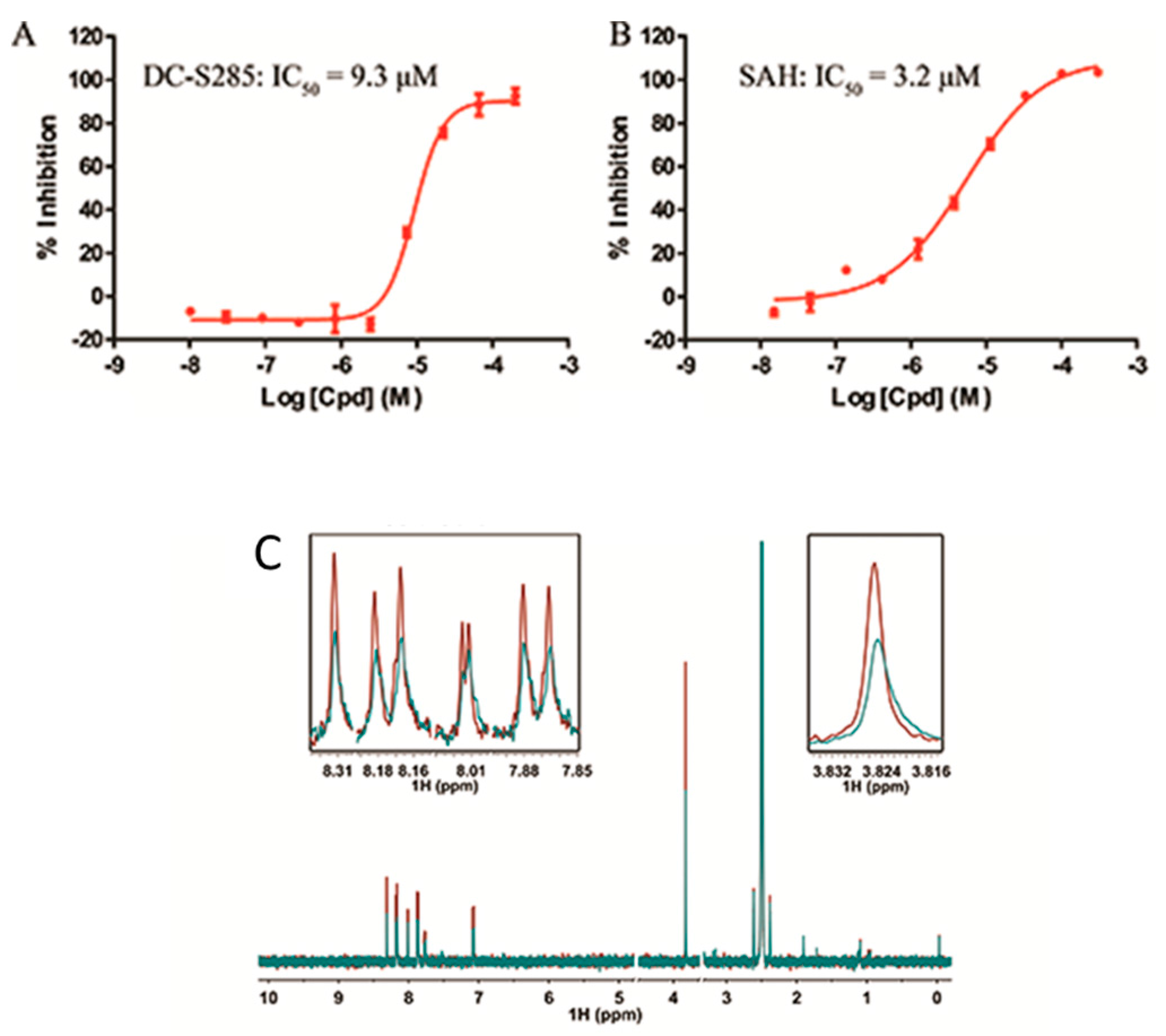
2.3. Validation of DC-S285′s Activity
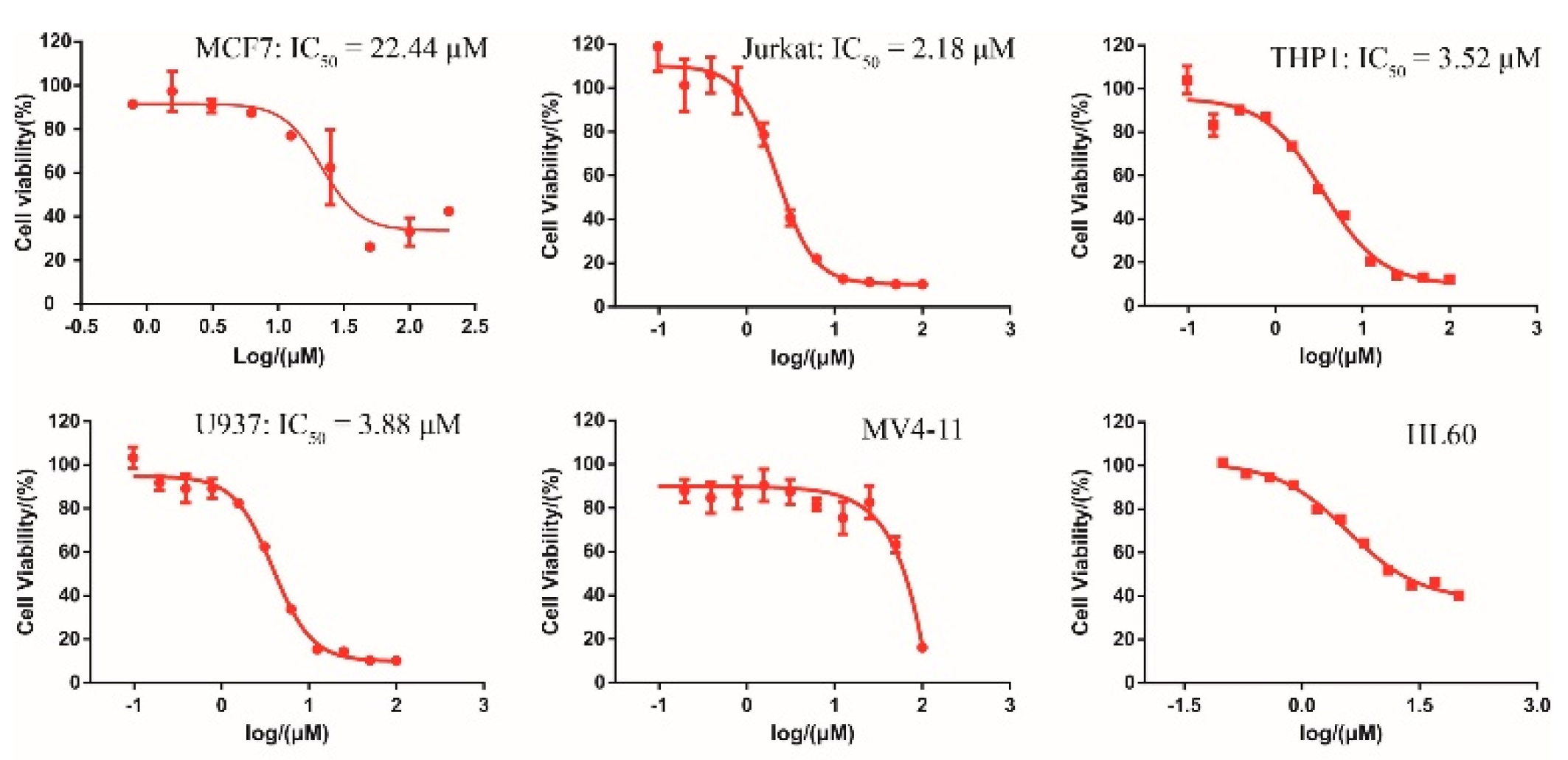
2.4. Cellular Activity of DC-S285
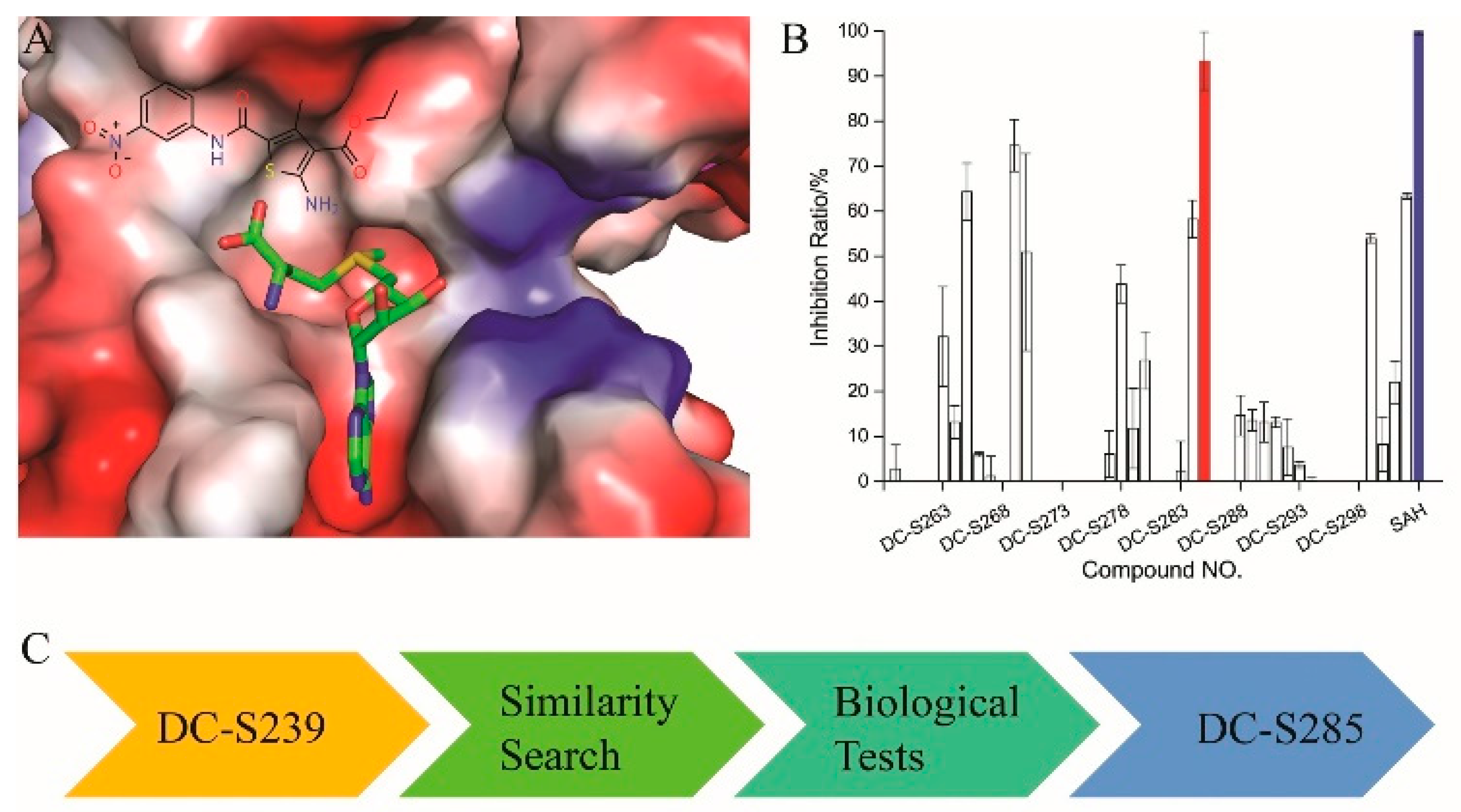
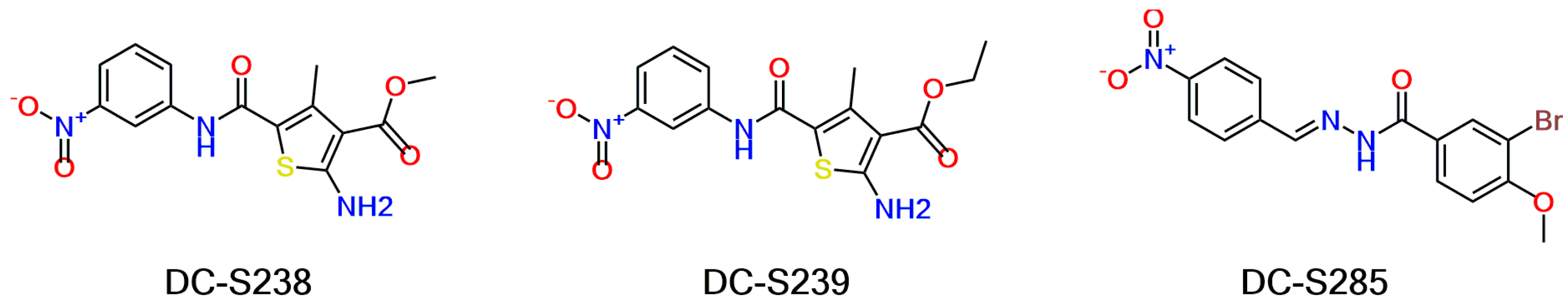
2.5. Similarity-Based Analog Searching and SAR Analysis
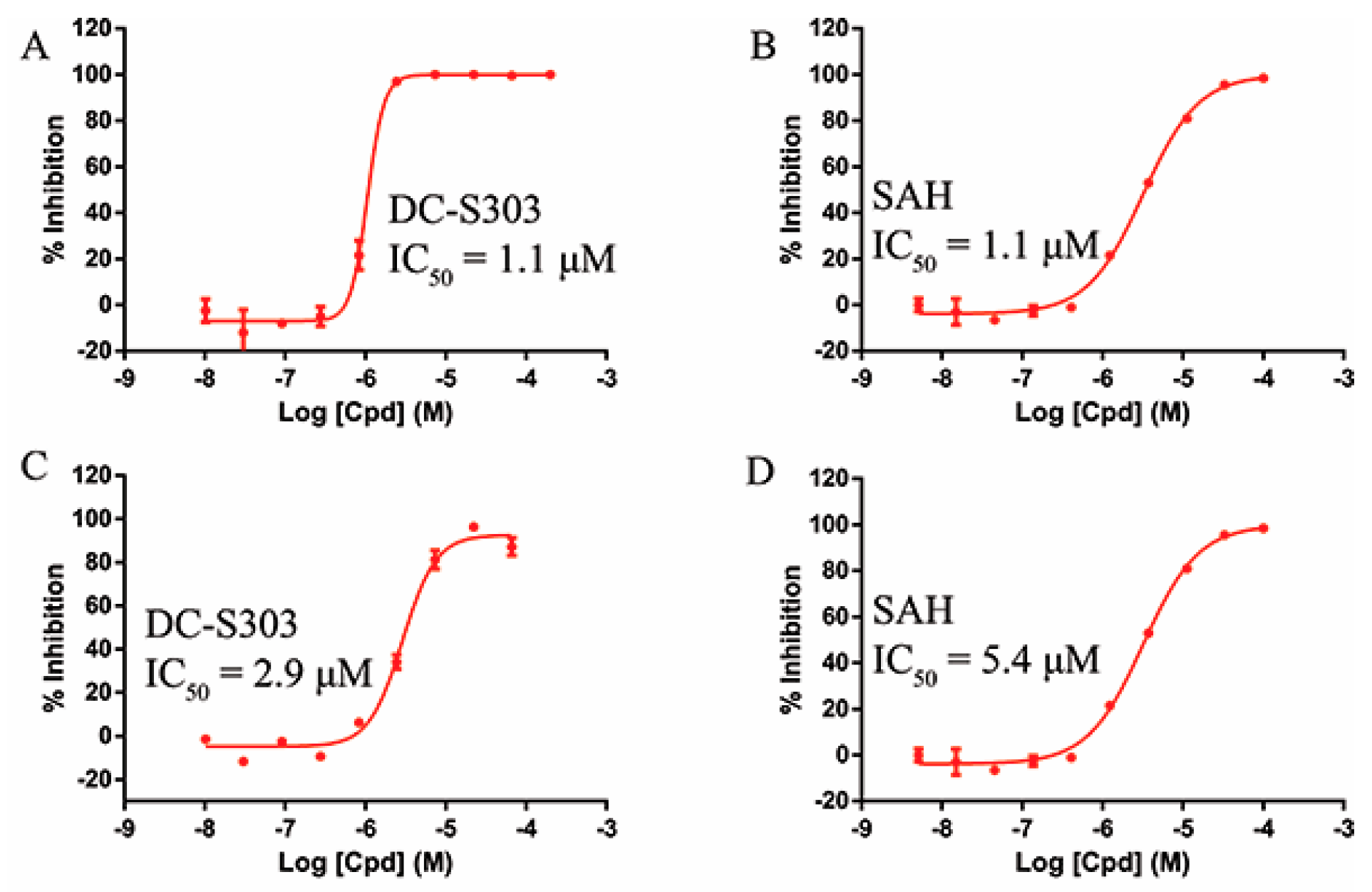
2.6. Selectivity of DC-S303
2.7. Binding Mode Prediction of DC-S303
3. Materials and Methods
3.1. Virtual Screening: Ligand Database Preparation
3.2. Virtual Screening: ProteinPreparation
3.3. Virtual Screening: 2D Molecular FingerprintBased Similarity Search
3.4. Virtual Screening: Scaffold Hopping Based Similarity Search
3.5. SET7 Inhibition Assays
3.6. Enzymatic Selectivity Assays
3.7. NMR Experiment
3.8. Cell Culture and Cell Viability Assay
4. Conclusions
Acknowledgments
Author Contribution
Conflicts of Interest
References
- Tajima, K.; Yae, T.; Javaid, S.; Tam, O.; Comaills, V.; Morris, R.; Wittner, B.S.; Liu, M.Z.; Engstrom, A.; Takahashi, F.; et al. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat. Commun. 2015, 6, 8257. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Hung, M.C.; Li, L.Y. EZH2: A pivotal regulator in controlling cell differentiation. Am. J. Transl. Res. 2012, 4, 364–375. [Google Scholar] [PubMed]
- Martens, M.B.; Frega, M.; Classen, J.; Epping, L.; Bijvank, E.; Benevento, M.; van Bokhoven, H.; Tiesinga, P.; Schubert, D.; Nadif Kasri, N. Euchromatin histone methyltransferase 1 regulates cortical neuronal network development. Sci. Rep. 2016, 6, 35756. [Google Scholar] [CrossRef] [PubMed]
- Herz, H.M.; Garruss, A.; Shilatifard, A. SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Jung, M. New lysine methyltransferase drug targets in cancer. Nat. Biotechnol. 2012, 30, 622–623. [Google Scholar] [CrossRef] [PubMed]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Issa, J.P.J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Tough, D.F.; Tak, P.P.; Tarakhovsky, A.; Prinjha, R.K. Epigenetic drug discovery: Breaking through the immune barrier. Nat. Rev. Drug Discov. 2016, 15, 835–853. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Zhang, Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011, 25, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; El-Osta, A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics 2013, 8, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhou, J.; Liu, X.; Lu, D.; Shen, C.; Du, Y.; Wei, F.Z.; Song, B.; Lu, X.; Yu, Y.; et al. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc. Natl. Acad. Sci. USA 2013, 110, 5516–5521. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.M.; Munro, S.; Kessler, B.; Oppermann, U.; La Thangue, N.B. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. Embo J. 2011, 30, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Carr, S.M.; La Thangue, N.B. Diversity within the pRb pathway: Is there a code of conduct? Oncogene 2012, 31, 4343–4352. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Q.; Lu, Q.; Jia, Z.; Chen, P.; Ma, K.; Wang, W.; Zhou, C. ISL-1 promotes pancreatic islet cell proliferation by forming an ISL-1/Set7/9/PDX-1 complex. Cell Cycle 2015, 14, 3820–3829. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Kim, W.K.; Oh, K.J.; Park, A.; Lee da, S.; Han, B.S.; Lee, S.C.; Bae, K.H. Methyltransferase and demethylase profiling studies during brown adipocyte differentiation. BMB Rep. 2016, 49, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Tuano, N.K.; Okabe, J.; Ziemann, M.; Cooper, M.E.; El-Osta, A. Set7 mediated interactions regulate transcriptional networks in embryonic stem cells. Nucleic Acids Res. 2016, 44, 9206–9217. [Google Scholar] [CrossRef] [PubMed]
- Castano, J.; Morera, C.; Sese, B.; Boue, S.; Bonet-Costa, C.; Marti, M.; Roque, A.; Jordan, A.; Barrero, M.J. SETD7 Regulates the Differentiation of Human Embryonic Stem Cells. PLoS ONE 2016, 11, e0149502. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.M.; La Thangue, N.B. Cell cycle control by a methylation-phosphorylation switch. Cell Cycle 2011, 10, 733–734. [Google Scholar] [CrossRef] [PubMed]
- Gaughan, L.; Stockley, J.; Wang, N.; McCracken, S.R.; Treumann, A.; Armstrong, K.; Shaheen, F.; Watt, K.; McEwan, I.J.; Wang, C.; et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 2011, 39, 1266–1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Geng, H.; Xue, C.H.; Beer, T.M.; Qian, D.Z. Functional regulation of hypoxia inducible factor-1 alpha by SET9 lysine methyltransferase. Bba-Mol. Cell Res. 2015, 1853, 881–891. [Google Scholar]
- Yang, J.; Huang, J.; Dasgupta, M.; Sears, N.; Miyagi, M.; Wang, B.; Chance, M.R.; Chen, X.; Du, Y.; Wang, Y.; et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21499–21504. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Suzuki, T.; Dohmae, N.; Hayami, S.; Unoki, M.; Yoshimatsu, M.; Toyokawa, G.; Takawa, M.; Chen, T.; Kurash, J.K.; et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011, 71, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Kim, E.; Koun, S.; Ham, H.J.; Rhee, M.; Kim, M.J.; Huh, T.L. Proper Activity of Histone H3 Lysine 4 (H3K4) Methyltransferase Is Required for Morphogenesis during Zebrafish Cardiogenesis. Mol. Cells 2015, 38, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H. p53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Toledo, F.; Wahl, G.M. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 2006, 6, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Borchers, C.; Tempst, P.; Zhang, Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 2001, 8, 1207–1217. [Google Scholar] [CrossRef]
- Nishioka, K.; Chuikov, S.; Sarma, K.; Erdjument-Bromage, H.; Allis, C.D.; Tempst, P.; Reinberg, D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Gene Dev. 2002, 16, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Dhayalan, A.; Kudithipudi, S.; Rathert, P.; Jeltsch, A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem. Biol. 2011, 18, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kassner, I.; Barandun, M.; Fey, M.; Rosenthal, F.; Hottiger, M.O. Crosstalk between SET7/9-dependent methylation and ARTD1-mediated ADP-ribosylation of histone H1.4. Epigenet. Chromatin 2013, 6, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassner, I.; Andersson, A.; Fey, M.; Tomas, M.; Ferrando-May, E.; Hottiger, M.O. SET7/9-dependent methylation of ARTD1 at K508 stimulates poly-ADP-ribose formation after oxidative stress. Open Biol. 2013, 3, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oppenheimer, H.; Kumar, A.; Meir, H.; Schwartz, I.; Zini, A.; Haze, A.; Kandel, L.; Mattan, Y.; Liebergall, M.; Dvir-Ginzberg, M. Set7/9 Impacts COL2A1 Expression Through Binding and Repression of SirT1 Histone Deacetylation. J. Bone Miner. Res. 2014, 29, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Esteve, P.O.; Chin, H.G.; Benner, J.; Feehery, G.R.; Samaranayake, M.; Horwitz, G.A.; Jacobsen, S.E.; Pradhan, S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA 2009, 106, 5076–5081. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hevi, S.; Kurash, J.K.; Lei, H.; Gay, F.; Bajko, J.; Su, H.; Sun, W.T.; Chang, H.; Xu, G.L.; et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009, 41, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Lezina, L.; Aksenova, V.; Ivanova, T.; Purmessur, N.; Antonov, A.V.; Tentler, D.; Fedorova, O.; Garabadgiu, A.V.; Talianidis, I.; Melino, G.; et al. KMTase Set7/9 is a critical regulator of E2F1 activity upon genotoxic stress. Cell Death Differ. 2014, 21, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Kontaki, H.; Talianidis, I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell 2010, 39, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, K.; Jia, D.; Kapoor-Vazirani, P.; Powell, D.R.; Collins, R.E.; Sharma, D.; Peng, J.M.; Cheng, X.D.; Vertino, P.M. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell 2008, 30, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Calnan, D.R.; Webb, A.E.; White, J.L.; Stowe, T.R.; Goswami, T.; Shi, X.; Espejo, A.; Bedford, M.T.; Gozani, O.; Gygi, S.P.; et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY) 2012, 4, 462–479. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniyan, N.; Ananthanarayanan, M.; Suchy, F.J. Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G937–G947. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.; Lin, J.; Zhou, L.; Song, Y.; Wei, B.; Luo, X.; Chen, Z.; Chen, Y.; Xiong, J.; et al. The transcription factor GATA1 and the histone methyltransferase SET7 interact to promote VEGF-mediated angiogenesis and tumor growth and predict clinical outcome of breast cancer. Oncotarget 2016, 7, 9859–9875. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wu, H.; Cheng, S.Y.; Gao, D.; Zhang, L.; Zhao, Y. Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals. eLife 2016, 5, e15690. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Z.; Xu, C.; Leng, X.; Cao, H.; Ouyang, G.; Xiao, W. Repression of hypoxia-inducible factor alpha signaling by Set7-mediated methylation. Nucleic Acids Res. 2015, 43, 5081–5098. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Kim, K.I. Regulation of HIF-1 alpha stability by lysine methylation. BMB Rep. 2016, 49, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Han, Q.; Nie, J.; Cao, X.; Chen, Z.; Yin, S.; Gao, Y.; Lin, F.; Zhou, X.; Xu, K.; et al. Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J. Biol. Chem. 2013, 288, 35093–35103. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Gao, D.; Zhang, W.; Liu, S.; Yang, S.; Li, X. Puerarin suppresses high glucose-induced MCP-1 expression via modulating histone methylation in cultured endothelial cells. Life Sci. 2015, 130, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Li, X.; Cui, P.; Li, Q.; Guo, Q.; Han, H.; Liu, S.; Sun, G. Involvement of Histone Lysine Methylation in p21 Gene Expression in Rat Kidney In Vivo and Rat Mesangial Cells In Vitro under Diabetic Conditions. J. Diabetes Res. 2016, 2016, 3853242. [Google Scholar] [CrossRef] [PubMed]
- Brasacchio, D.; Okabe, J.; Tikellis, C.; Balcerczyk, A.; George, P.; Baker, E.K.; Calkin, A.C.; Brownlee, M.; Cooper, M.E.; El-Osta, A. Hyperglycemia Induces a Dynamic Cooperativity of Histone Methylase and Demethylase Enzymes Associated with Gene-Activating Epigenetic Marks That Coexist on the Lysine Tail. Diabetes 2009, 58, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Ea, C.K.; Baltimore, D. Regulation of NF-kappa B activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. USA 2009, 106, 18972–18977. [Google Scholar] [CrossRef] [PubMed]
- Masatsugu, T.; Yamamoto, K. Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochem Biophys. Res. Commun. 2009, 381, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Spaeth, J.M.; Walker, E.M.; Stein, R. Impact of Pdx1-associated chromatin modifiers on islet beta-cells. Diabetes Obes. Metab. 2016, 18 (Suppl. 1), 123–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shi, Q.; Guo, T.; Yang, Z.; Jia, Z.; Chen, P.; Zhou, C. PDX1 and ISL1 differentially coordinate with epigenetic modifications to regulate insulin gene expression in varied glucose concentrations. Mol. Cell. Endocrinol. 2016, 428, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Maganti, A.V.; Maier, B.; Tersey, S.A.; Sampley, M.L.; Mosley, A.L.; Ozcan, S.; Pachaiyappan, B.; Woster, P.M.; Hunter, C.S.; Stein, R.; et al. Transcriptional Activity of the Islet beta Cell Factor Pdx1 Is Augmented by Lysine Methylation Catalyzed by the Methyltransferase Set7/9. J. Biol. Chem. 2015, 290, 9812–9822. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.D.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1 alpha. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Huq, M.D.; Tsai, N.P.; Khan, S.A.; Wei, L.N. Lysine trimethylation of retinoic acid receptor-alpha: A novel means to regulate receptor function. Mol. Cell. Proteom. 2007, 6, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, Y.; Gao, Y.; Geng, P.; Lu, Y.; Liu, X.; Yao, R.; Hou, P.; Liu, D.; Lu, J.; et al. JMJD3 promotes SAHF formation in senescent WI38 cells by triggering an interplay between demethylation and phosphorylation of RB protein. Cell Death Differ. 2015, 22, 1630–1640. [Google Scholar] [CrossRef] [PubMed]
- He, S.Y.; Owen, D.R.; Jelinsky, S.A.; Lin, L.L. Lysine Methyltransferase SETD7 (SET7/9) Regulates ROS Signaling through mitochondria and NFE2L2/ARE pathway. Sci. Rep.-UK 2015, 5, 14368. [Google Scholar] [CrossRef] [PubMed]
- Elkouris, M.; Kontaki, H.; Stavropoulos, A.; Antonoglou, A.; Nikolaou, K.C.; Samiotaki, M.; Szantai, E.; Saviolaki, D.; Brown, P.J.; Sideras, P.; et al. SET9-Mediated Regulation of TGF-beta Signaling Links Protein Methylation to Pulmonary Fibrosis. Cell Rep. 2016, 15, 2733–2744. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, E.; Fedorova, O.; Shuvalov, O.; Daks, A.; Petukhov, A.; Barlev, N. The role of methyltransferase Set7/9 interaction with RNA-binding protein Sam68. Int. J. Mol. Med. 2015, 36, S46. [Google Scholar]
- Liu, X.Y.; Wang, D.L.; Zhao, Y.; Tu, B.; Zheng, Z.X.; Wang, L.N.; Wang, H.Y.; Gu, W.; Roeder, R.G.; Zhu, W.G. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc. Natl. Acad. Sci. USA 2011, 108, 1925–1930. [Google Scholar] [CrossRef] [PubMed]
- Albacker, C.E.; Storer, N.Y.; Langdon, E.M.; DiBiase, A.; Zhou, Y.; Langenau, D.M.; Zon, L.I. The Histone Methyltransferase SUV39H1 Suppresses Embryonal Rhabdomyosarcoma Formation in Zebrafish. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couture, J.F.; Collazo, E.; Hauk, G.; Trievel, R.C. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 2006, 13, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Kouskouti, A.; Scheer, E.; Staub, A.; Tora, L.; Talianidis, I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell 2004, 14, 175–182. [Google Scholar] [CrossRef]
- Pagans, S.; Kauder, S.E.; Kaehlcke, K.; Sakane, N.; Schroeder, S.; Dormeyer, W.; Trievel, R.C.; Verdin, E.; Schnolzer, M.; Ott, M. The Cellular Lysine Methyltransferase Set7/9-KMT7 Binds HIV-1 TAR RNA, Monomethylates the Viral Transactivator Tat, and Enhances HIV Transcription. Cell Host Microbe 2010, 7, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.Y.; Li, X.X.; Han, H.B.; Li, C.Y.; Liu, S.J.; Gao, W.H.; Sun, G.D. Histone Lysine Methylation in TGF-beta 1 Mediated p21 Gene Expression in Rat Mesangial Cells. Biomed. Res. Int. 2016, 2016, 6927234. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Doi, S.; Nakashima, A.; Irifuku, T.; Yamada, K.; Kokoroishi, K.; Ueno, T.; Doi, T.; Hida, E.; Arihiro, K.; et al. Inhibition of SET Domain-Containing Lysine Methyltransferase 7/9 Ameliorates Renal Fibrosis. J. Am. Soc. Nephrol. 2016, 27, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Madapura, M.P.; Bhat, U.A.; Rao, M.R.S. Mapping of Post-translational Modifications of Transition Proteins, TP1 and TP2, and Identification of Protein Arginine Methyltransferase 4 and Lysine Methyltransferase 7 as Methyltransferase for TP2. J. Biol. Chem. 2015, 290, 12101–12122. [Google Scholar] [CrossRef] [PubMed]
- Oudhoff, M.J.; Braam, M.J.S.; Freeman, S.A.; Wong, D.; Rattray, D.G.; Wang, J.; Antignano, F.; Snyder, K.; Refaeli, I.; Hughes, M.R.; et al. SETD7 Controls Intestinal Regeneration and Tumorigenesis by Regulating Wnt/beta-Catenin and Hippo/YAP Signaling. Dev. Cell 2016, 37, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Wu, X.N.; Shi, T.T.; Xu, H.T.; Yi, J.; Shen, H.F.; Huang, M.F.; Shu, X.Y.; Wang, F.F.; Peng, B.L.; et al. Regulation of Transcription Factor Yin Yang 1 by SET7/9-mediated Lysine Methylation. Sci. Rep.-UK 2016, 6, 21718. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liang, G.; Wu, X.; Wang, S.; Zhang, P.; Su, Y.; Yin, H.; Tan, Y.; Zhang, J.; Lu, Q. Abnormal epigenetic modifications in peripheral blood mononuclear cells from patients with alopecia areata. Br. J. Dermatol. 2012, 166, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.C.; Zhang, J.L.; Tian, T.; Fu, X.; Wang, W.J.; Li, S.N.; Shi, T.T.; Suo, A.L.; Ruan, Z.P.; Guo, H.; et al. SET7/9 inhibits oncogenic activities through regulation of Gli-1 expression in breast cancer. Tumor Biol. 2016, 37, 9311–9322. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Yang, S.S.; Hu, J.W.; Yu, C.Q.; He, M.X.; Cai, Z.L. Increased Expression of SETD7 Promotes Cell Proliferation by Regulating Cell Cycle and Indicates Poor Prognosis in Hepatocellular Carcinoma. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Takeuchi, T.; Nagayama, T.; Furihata, M. T-Cadherin Modulates Tumor-Associated Molecules in Gallbladder Cancer Cells. Cancer Investig. 2010, 28, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Koda, Y.; Byeon, S.J.; Shimada, S.; Nishikawaji, T.; Sakamoto, A.; Chen, Y.X.; Kojima, K.; Kawano, T.; Eishi, Y.; et al. Reduced expression of SET7/9, a histone mono-methyltransferase, is associated with gastric cancer progression. Oncotarget 2016, 7, 3966–3983. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Nam, H.J.; Lee, J.; Park, D.Y.; Kim, C.; Yu, Y.S.; Kim, D.; Park, S.W.; Bhin, J.; Hwang, D.; et al. Methylation-dependent regulation of HIF-1 alpha stability restricts retinal and tumour angiogenesis. Nat. Commun. 2016, 7, 10347. [Google Scholar] [CrossRef] [PubMed]
- Lezina, L.; Aksenova, V.; Fedorova, O.; Malikova, D.; Shuvalov, O.; Antonov, A.V.; Tentler, D.; Garabadgiu, A.V.; Melino, G.; Barlev, N.A. KMT Set7/9 affects genotoxic stress response via the Mdm2 axis. Oncotarget 2015, 6, 25843–25855. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.C.; Wang, D.L.; Liu, X.Y.; Gu, B.; Du, Y.P.; Wei, F.Z.; Cao, L.L.; Song, B.Y.; Lu, X.P.; Yang, Q.Y.; et al. SET7/9 regulates cancer cell proliferation by influencing beta-catenin stability. Faseb J. 2015, 29, 4313–4323. [Google Scholar] [CrossRef] [PubMed]
- Greissel, A.; Culmes, M.; Burgkart, R.; Zimmermann, A.; Eckstein, H.H.; Zernecke, A.; Pelisek, J. Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc. Pathol. 2016, 25, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Reddy, M.A.; Deshpande, S.; Jia, Y.; Park, J.T.; Lanting, L.L.; Jin, W.; Kato, M.; Xu, Z.G.; Das, S.; et al. Epigenetic Histone Modifications Involved in Profibrotic Gene Regulation by 12/15-Lipoxygenase and Its Oxidized Lipid Products in Diabetic Nephropathy. Antioxid. Redox. Signal. 2016, 24, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Paneni, F.; Costantino, S.; Battista, R.; Castello, L.; Capretti, G.; Chiandotto, S.; Scavone, G.; Villano, A.; Pitocco, D.; Lanza, G.; Volpe, M.; et al. Adverse epigenetic signatures by histone methyltransferase Set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circ. Cardiovasc. Genet. 2015, 8, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Okabe, J.; Orlowski, C.; Balcerczyk, A.; Tikellis, C.; Thomas, M.C.; Cooper, M.E.; El-Osta, A. Distinguishing Hyperglycemic Changes by Set7 in Vascular Endothelial Cells. Circ. Res. 2012, 110, 1067–1121. [Google Scholar] [CrossRef] [PubMed]
- Goru, S.K.; Kadakol, A.; Pandey, A.; Malek, V.; Sharma, N.; Gaikwad, A.B. Histone H2AK119 and H2BK120 mono-ubiquitination modulate SET7/9 and SUV39H1 in type 1 diabetes-induced renal fibrosis. Biochem. J. 2016, 473, 3937–3949. [Google Scholar] [CrossRef] [PubMed]
- Costantino, S.; Paneni, F.; Virdis, A.; Volpe, M.; Taddei, S.; Cosentino, F. Chromatin changes by methyltransferase Set7 induce inflammatory adhesion molecules and endothelial dysfunction in small visceral arteries from insulin resistant subjects. Eur. Heart J. 2015, 36, 112–113. [Google Scholar]
- Ciccarelli, M.; Vastolo, V.; Albano, L.; Lecce, M.; Cabaro, S.; Liotti, A.; Longo, M.; Oriente, F.; Russo, G.L.; Macchia, P.E.; et al. Glucose-induced expression of the homeotic transcription factor Prep1 is associated with histone post-translational modifications in skeletal muscle. Diabetologia 2016, 59, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Al-Haddad, R.; Karnib, N.; Assaad, R.A.; Bilen, Y.; Emmanuel, N.; Ghanem, A.; Younes, J.; Zibara, V.; Stephan, J.S.; Sleiman, S.F. Epigenetic changes in diabetes. Neurosci. Lett. 2016, 625, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Son, M.J.; Kim, W.K.; Park, A.; Oh, K.J.; Kim, J.H.; Han, B.S.; Kim, I.C.; Chi, S.W.; Park, S.G.; Lee, S.C.; et al. Set7/9, a methyltransferase, regulates the thermogenic program during brown adipocyte differentiation through the modulation of p53 acetylation. Mol. Cell. Endocrinol. 2016, 431, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xie, M.; Shi, Y.; Luo, B.H.; Gong, G.H.; Li, J.N.; Wang, J.P.; Zhao, W.J.; Zi, Y.; Wu, X.Y.; et al. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol. Rep. 2015, 34, 111–120. [Google Scholar] [CrossRef] [PubMed]
- D’Adamo, C.R.; D’Urso, A.; Ryan, K.A.; Yerges-Armstrong, L.M.; Semba, R.D.; Steinle, N.I.; Mitchell, B.D.; Shuldiner, A.R.; McArdle, P.F. A Common Variant in the SETD7 Gene Predicts Serum Lycopene Concentrations. Nutrients 2016, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Wan, Y.S.; Wang, J.; Zhao, P.; Yuan, Y.; Wang, L.; She, Y.L.; Broering, R.; Lu, M.J.; Ye, L.B.; et al. Set7 Facilitates Hepatitis C Virus Replication via Enzymatic Activity-Dependent Attenuation of the IFN-Related Pathway. J. Immunol. 2015, 194, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Ramage, H.; Boehm, D.; Dirk, L.M.A.; Sakane, N.; Hanada, K.; Pagans, S.; Kaehlcke, K.; Aull, K.; Weinberger, L.; et al. The HIV-1 Tat Protein Is Monomethylated at Lysine 71 by the Lysine Methyltransferase KMT7. J. Biol. Chem. 2016, 291, 16240–16248. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.H.; Peng, K.L.; Jhang, H.C.; Lin, C.H.; Wu, S.Y.; Chiang, C.M.; Lee, S.C.; Yu, W.C.Y.; Juan, L.J. The HPV E6 oncoprotein targets histone methyltransferases for modulating specific gene transcription. Oncogene 2012, 31, 2335–2349. [Google Scholar] [CrossRef] [PubMed]
- Barsyte-Lovejoy, D.; Li, F.L.; Oudhoff, M.J.; Tatlock, J.H.; Dong, A.P.; Zeng, H.; Wu, H.; Freeman, S.A.; Schapira, M.; Senisterra, G.A.; et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. USA 2014, 111, 12853–12858. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.W.; Cheng, S.F.; Ding, H.; Liu, S.; Liu, Y.; Zhu, K.K.; Chen, S.J.; Lu, J.Y.; Xie, Y.Q.; Li, L.J.; et al. Selective Histone Methyltransferase SET7 Inhibitors by Pharmacophore- and Docking-Based Virtual Screening. J. Med. Chem. 2015, 58, 8166–8181. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.Y.; Cai, C.Q.; Liu, X.F.; Ku, X.; Jiang, H.L.; Gao, D.Q.; Li, H.L. ChemMapper: A versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method. Bioinformatics 2013, 29, 1827–1829. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.P.; Hersey, A.; Montanari, D.; Overington, J. Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat. Rev. Drug Discov. 2011, 10, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Van de Waterbeemd, H.; Gifford, E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Whitty, A. Growing PAINS in academic drug discovery. Future Med. Chem. 2011, 3, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.; Walters, M.A. Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B. Observations on screening-based research and some concerning trends in the literature. Future Med. Chem. 2010, 2, 1529–1546. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pK (a) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

| Substrates | Functions | References | |
|---|---|---|---|
| Histone | H3K4 | transcriptional activation | [27,28] |
| H2A | [29] | ||
| H2BK15 | [29] | ||
| H1.4 | [30] | ||
| Non-histone | ARTD1 | stimulating poly-ADP-ribose formation after oxidative stress | [31] |
| COL2A1 | morphology-dependent COL2A1 gene transactivation | [32] | |
| DNMT1 | protein stability regulation of DNMT1 (destabilization) | [33,34] | |
| E2F1 | regulation of E2F1 stabilization co-activator in response to DNA damage | [35,36] | |
| ERα | protein stability regulation of ERα (stabilization) and enhancing transcriptional activity | [20,37] | |
| FoxO3 | protein stability regulation of FoxO3 (destabilization) | [38] | |
| FXR | transcriptional activation of FXR-target genes | [39] | |
| GATA1 | required for GATA1-induced breast tumour angiogenesis and growth in nude mice; poor prognostic factors in breast cancer | [40] | |
| Gli3 | activation of Sonic Hedgehog pathway in mammals | [41] | |
| HIF-1α | promoting HIF-1α protein stability in hypoxia and enhancing HIF-1 mediated glycolytic gene transcription | [21] | |
| HIF-1α/2α | negatively regulation HIF-α transcriptional activity and HIF-1-mediated glucose homeostasis | [42,43] | |
| IFITM3 | negatively affected IFITM3 antiviral activity | [44] | |
| MCP-1 | regulation of MCP-1 mRNA expression | [45] | |
| MYPT1 | protein stability regulation of MYPT1 (stabilization) | [23] | |
| p21 | [46] | ||
| p53 | |||
| p65 (RelA) | regulation of NF-κB activity | [11,47,48] | |
| PCAF | [49] | ||
| PDX1 | maintenance of Pdx1 activity and β cell function; control insulin gene expression based on glucose concentration | [50,51,52] | |
| PCG-1α | [53] | ||
| pRb | cell cycle arrest | [13,14] | |
| RARα | [54] | ||
| RB | promotes cell cycle progression | [55] | |
| NFE2L2 | negatively regulates the expression of NFE2L2 and its downstream genes | [56] | |
| Smad7 | [57,58] | ||
| SIRT1 | inducing the dissociation of SIRT1 from p53 and increasing p52 activity | [59] | |
| STAT3 | negatively regulation of protein stability and transactivation activity | [22] | |
| SUV39H1 | gene instability and cell proliferation inhibition | [12,60] | |
| TAF7 | RNA polymerase ii-dependent transcription coactivator | [61] | |
| TAF10 | RNA polymerase ii-dependent transcription coactivator | [62] | |
| TAT | enhancing HIV transcription | [63] | |
| TGF-β1 | transcriptional activation of fibrotic genes | [64,65] | |
| TP2 | elongating to condensing spermatids | [66] | |
| YAP | control YAP subcellular localization and function | [67] | |
| YY1 | regulation of YY1 DNA-binding activity | [68] | |
| AKA6 | unknown | [29] | |
| CENPC | unknown | [29] | |
| MeCP2 | unknown | [29] | |
| MINT | unknown | [29] | |
| PPARBP, | unknown | [29] | |
| ZDH8 | unknown | [29] | |
| Cullin1 | unknown | [29] | |
| IRF1/2 | unknown | [29] | |

| No. | R1 | R2 | R3 | Inhibition Ratio at 100 μM/% | IC50 (μM) |
|---|---|---|---|---|---|
| DC-S303 |  |  (A) |  | 99 | 1.1 |
| DC-S304 |  | 44 | |||
| DC-S305 |  | −5 | |||
| DC-S306 |  | 94 | 20 | ||
| DC-S307 |  | 8 | |||
| DC-S308 |  | 7 | |||
| DC-S309 |  | −5 | |||
| DC-S310 |  | −6 | |||
| DC-S311 |  | 97 | 13 | ||
| DC-S312 |  | −3 | |||
| DC-S313 |  | −6 | |||
| DC-S314 |  |  (B) |  | 77 | 46 |
| DC-S315 |  | 46 | |||
| DC-S316 |  | 37 | |||
| DC-S317 |  | 29 | |||
| DC-S318 |  | 17 | |||
| DC-S319 |  | 14 | |||
| DC-S320 |  | 11 | |||
| DC-S321 |  | 9 | |||
| DC-S334 |  |  (C) |  | 96 | 9.9 |
| DC-S335 |  | 12 | IC50 (μM) | ||
| DC-S336 |  | 49 | 1.1 | ||
| DC-S337 |  | 1 | |||
| DC-S338 |  | 1 | |||
| DC-S339 |  | −6 | 20 | ||
| DC-S340 |  | −6 | |||
| DC-S341 |  | −10 | |||
| DC-S342 |  | −13 | |||
| DC-S343 |  |  (D) |  | 1 | |
| DC-S344 |  | 32 | 13 | ||
| DC-S345 |  | 29 | |||
| DC-S346 |  | 21 | |||
| DC-S347 |  | 10 | |||
| DC-S348 |  | −11 | |||
| DC-S349 |  | −1 | |||
| DC-S350 |  | 6 | |||
| DC-S351 |  | 96 | 3.4 | ||
| DC-S352 |  | 39 | |||
| DC-S353 |  | 35 | |||
| DC-S354 |  | 35 | |||
| DC-S355 |  | 20 | |||
| DC-S356 |  | 17 | |||
| DC-S357 |  | 13 | |||
| DC-S358 |  | 6 | |||
| DC-S359 |  | 5 | |||
| DC-S360 |  | 4 | |||
| DC-S361 |  | −1 | |||
| DC-S362 |  | −4 | |||
| DC-S363 |  | −15 | |||
| DC-S364 |  | 54 | 3.7 | ||
| DC-S365 |  |  (E) |  | 10 | |
| DC-S366 |  | 9 | |||
| DC-S367 |  | −1 |
| Compound No. | Target | Inhibition Ratio at 100 μM/% |
|---|---|---|
| DC-S303 | SETD7 | 90.51 |
| SETD1B | 27.12 | |
| SETD8 | 55.23 | |
| G9a | 52.56 | |
| SMYD2 | 24.55 | |
| EZH2 | 47.88 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, H.; Lu, W.C.; Hu, J.C.; Liu, Y.-C.; Zhang, C.H.; Lian, F.L.; Zhang, N.X.; Meng, F.W.; Luo, C.; Chen, K.X. Identification and Characterizations of Novel, Selective Histone Methyltransferase SET7 Inhibitors by Scaffold Hopping- and 2D-Molecular Fingerprint-Based Similarity Search. Molecules 2018, 23, 567. https://doi.org/10.3390/molecules23030567
Ding H, Lu WC, Hu JC, Liu Y-C, Zhang CH, Lian FL, Zhang NX, Meng FW, Luo C, Chen KX. Identification and Characterizations of Novel, Selective Histone Methyltransferase SET7 Inhibitors by Scaffold Hopping- and 2D-Molecular Fingerprint-Based Similarity Search. Molecules. 2018; 23(3):567. https://doi.org/10.3390/molecules23030567
Chicago/Turabian StyleDing, Hong, Wen Chao Lu, Jun Chi Hu, Yu-Chih Liu, Chen Hua Zhang, Fu Lin Lian, Nai Xia Zhang, Fan Wang Meng, Cheng Luo, and Kai Xian Chen. 2018. "Identification and Characterizations of Novel, Selective Histone Methyltransferase SET7 Inhibitors by Scaffold Hopping- and 2D-Molecular Fingerprint-Based Similarity Search" Molecules 23, no. 3: 567. https://doi.org/10.3390/molecules23030567