Synthesis, Molecular Docking, and Antimycotic Evaluation of Some 3-Acyl Imidazo[1,2-a]pyrimidines
Abstract
:1. Introduction
2. Results
2.1. Synthesis
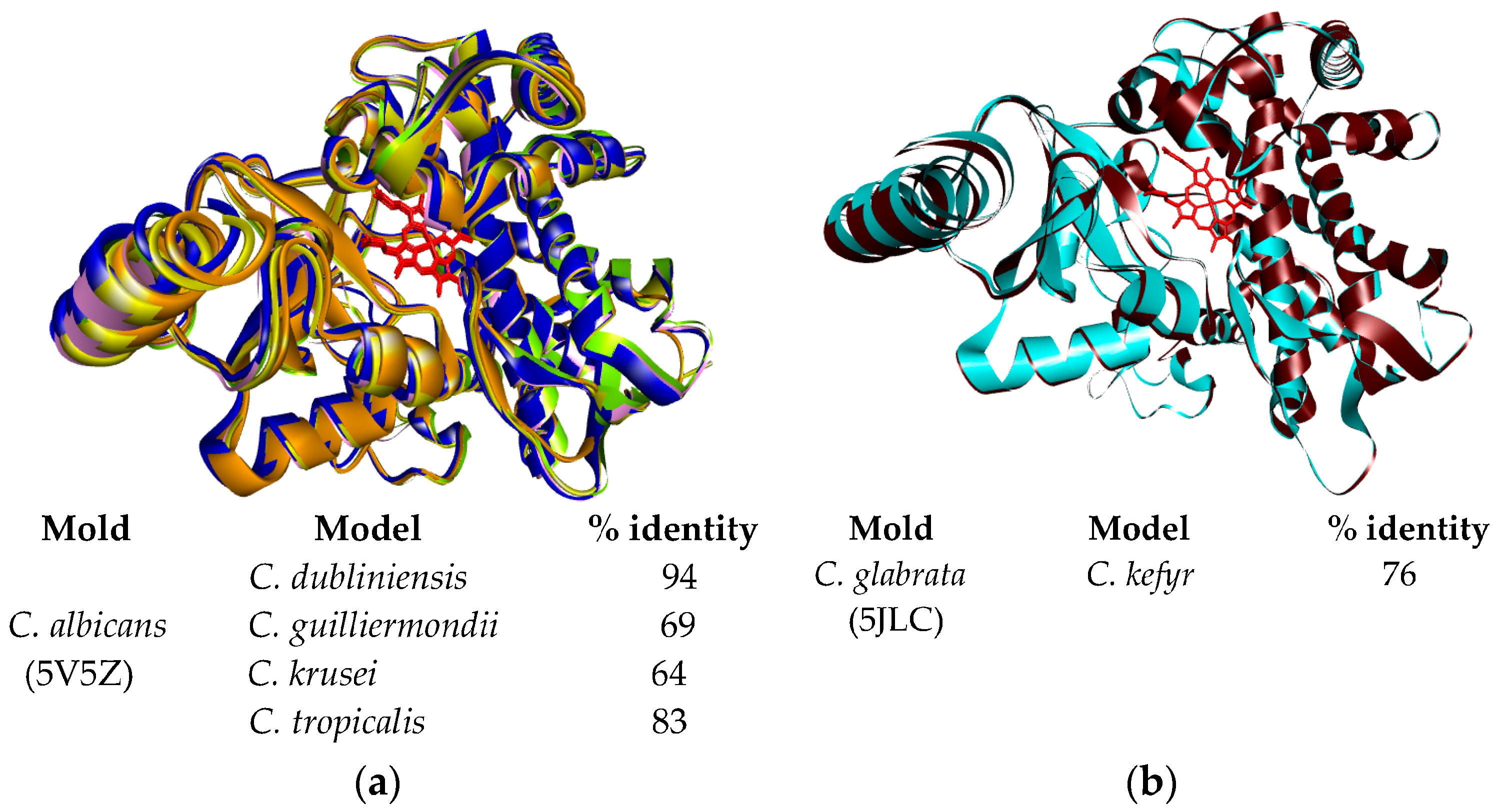
2.2. Modeling CYP51 from Candida spp.
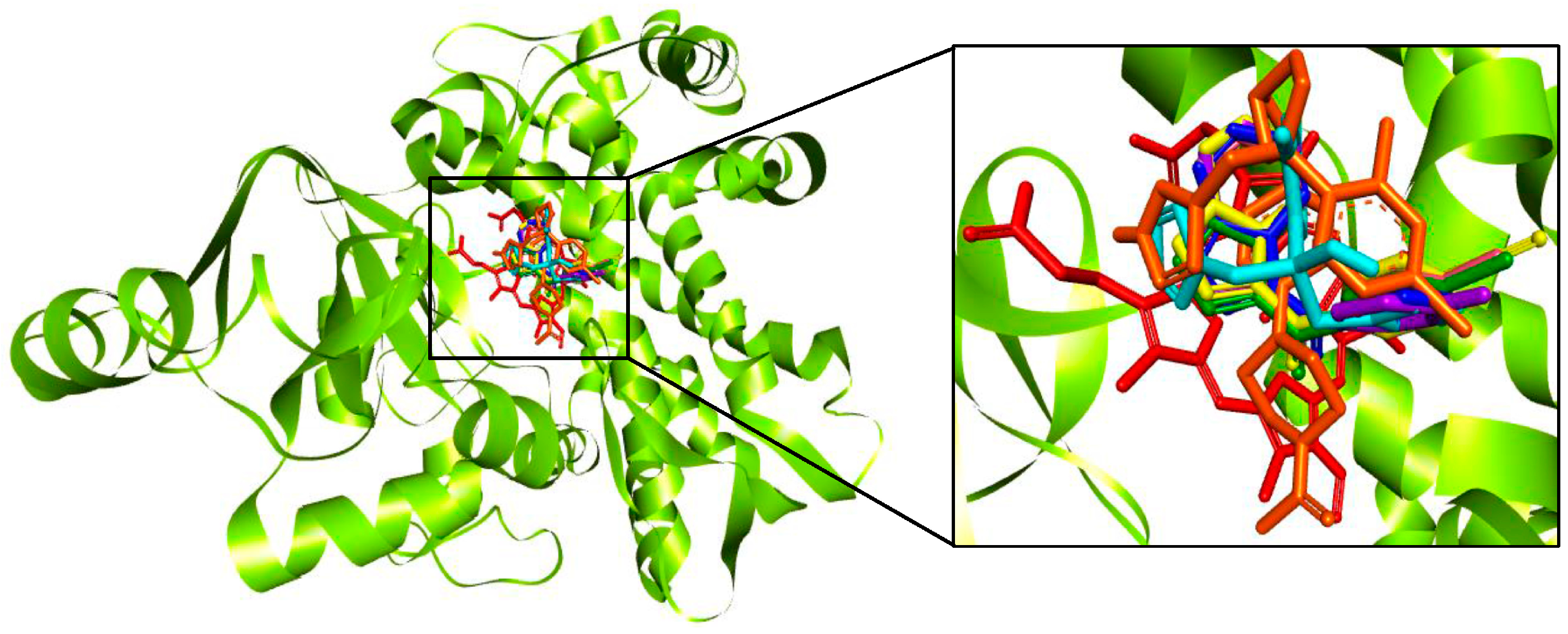
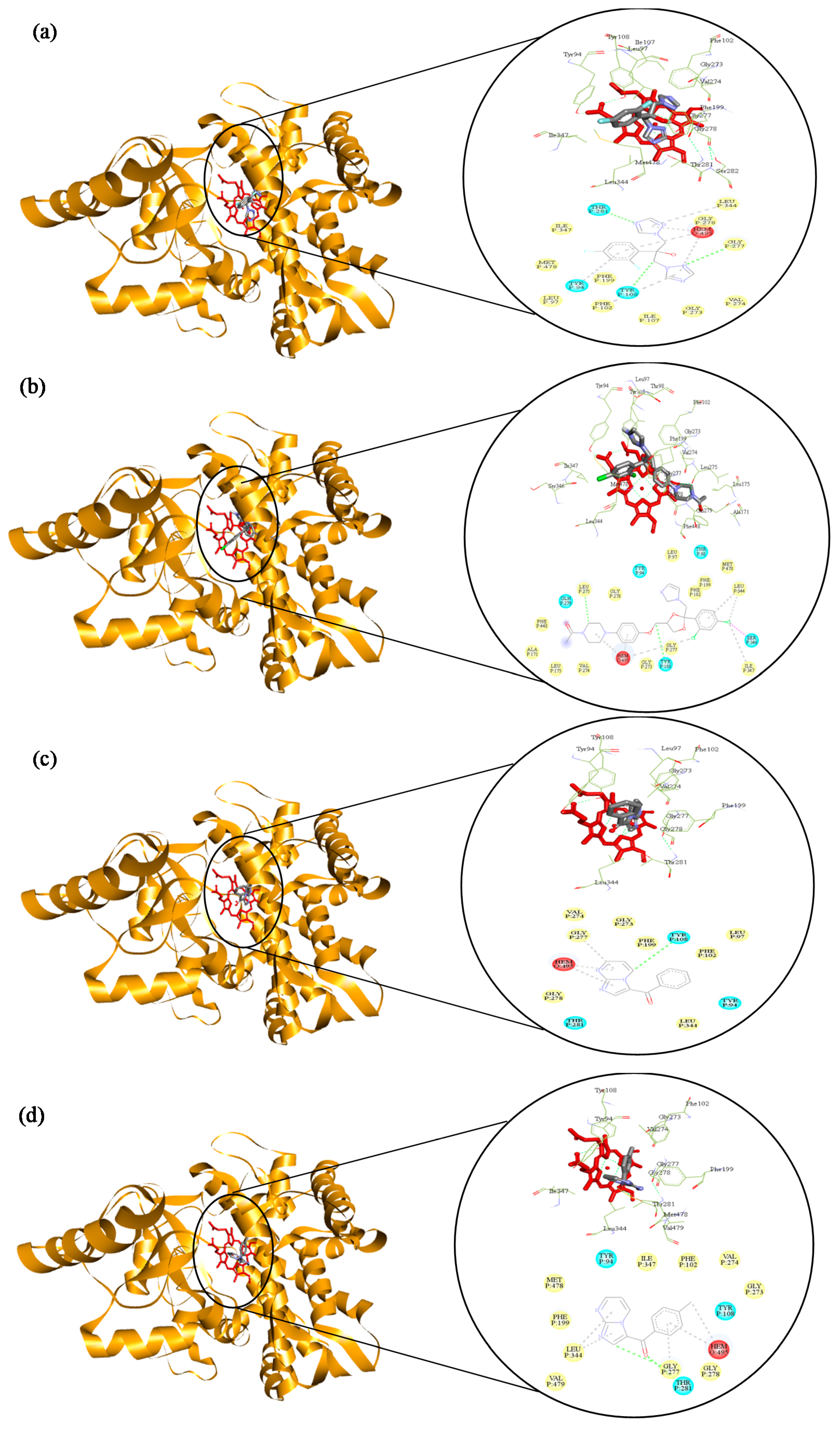
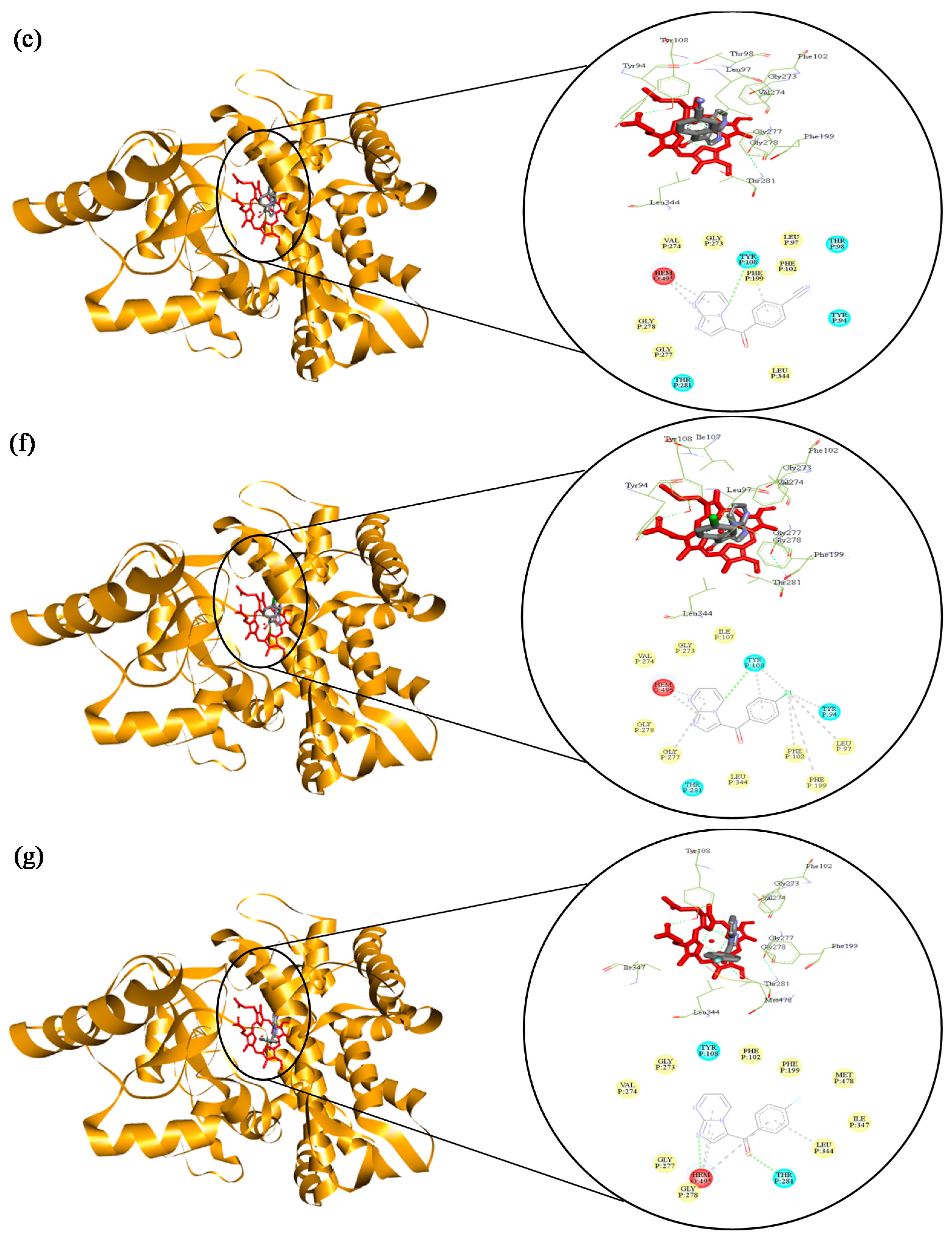
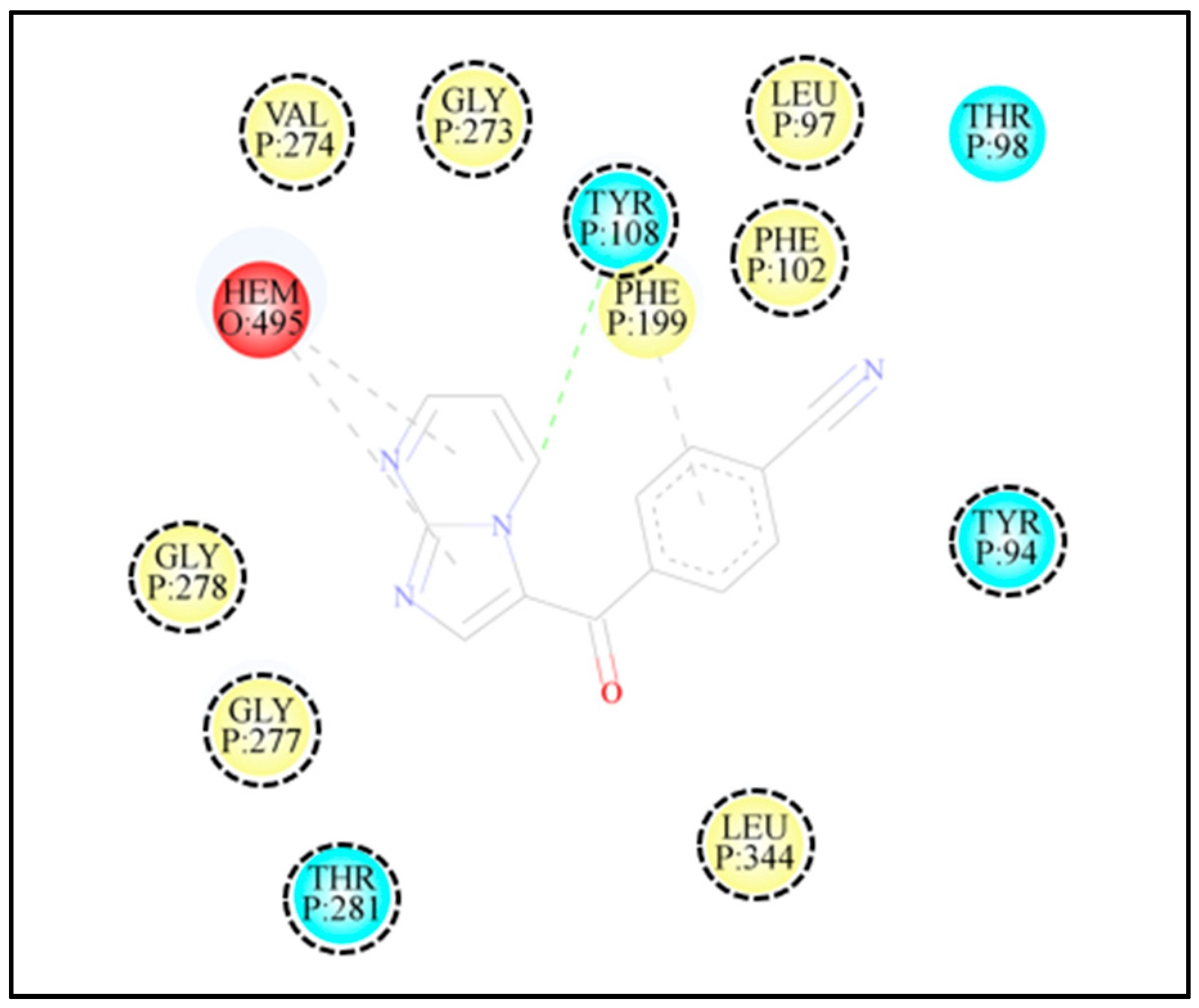
2.3. Molecular Docking of 3-Benzoyl Imidazo[1,2-a]pyrimidines in the Active Site of CYP51 of Candida spp.
2.4. Susceptibility of Candida spp. to 3-Benzoylimidazo[1,2-a]pyrimidines
3. Discussion
4. Materials and Methods
4.1. Chemicals and Instruments
4.2. Synthesis of Imidazo[1,2-a]pyrimidin-3-yl(phenyl)methanones (3a–j)
4.3. Microorganisms and Compounds Tested
4.4. Homology Modeling
4.5. Molecular Docking Studies
4.6. Antifungal Activity Tests
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Aeluri, R.; Alla, M.; Polepalli, S.; Jain, N. Synthesis and antiproliferative activity of imidazo[1,2-a]Pyrimidine Mannich Bases. Eur. J. Med. Chem. 2015, 100, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ai, J.; Zhang, D.; Peng, X.; Chen, X.; Gao, Z.; Su, Y.; Zhu, W.; Ji, Y.; Chen, X.; et al. Design, synthesis, and biological evaluation of novel imidazo[1,2-a]pyridine derivatives as potent c-Met Inhibitor. ACS Med. Chem. Lett. 2015, 6, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Reddy, J.S.; Ramaiah, M.J.; Dastagiri, D.; Bharathi, E.V.; Sagar, V.P.; Pushpavalli, S.N.; Ray, P.; Pal-Bhadra, M. Design, synthesis and biological evaluation of imidazopyridine/pyrimidine-chalcone derivatives as potential anticancer agents. Med. Chem. Commun. 2010, 1, 355–360. [Google Scholar] [CrossRef]
- Meng, W.; Brigance, R.P.; Chao, H.J.; Fura, A.; Harrity, T.; Marcinkeviciene, J.; O’Connor, S.P.; Tamura, J.K.; Xie, D.; Zhang, Y.; et al. Discovery of 6-(Aminomethyl)-5-(2,4-dichlorophenyl)-7-methylimidazo[1,2-a]pyrimidine-2-carboxamides as Potent, Selective Dipeptidyl Peptidase-4 (DPP4) Inhibitors. J. Med. Chem. 2010, 53, 5620–5628. [Google Scholar] [CrossRef] [PubMed]
- Moraski, G.C.; Markley, L.D.; Chang, M.; Cho, S.; Franzblau, S.G.; Hwang, C.H.; Boshoff, H.; Miller, M.J. Generation and exploration of new classes of antitubercular agents: The optimization of oxazolines, oxazoles, thiazolines, thiazoles to imidazo[1,2-a]pyridines and isomeric 5,6-fused scaffolds. Bioorg. Med. Chem. 2012, 20, 2214–2220. [Google Scholar] [CrossRef] [PubMed]
- Gueiffier, A.; Lhassani, M.; Elhakmaoui, A.; Snoeck, R.; Andrei, G.; Chavignon, O.; Teulade, J.C.; Kerbal, A.; Essassi, E.M.; Debouzy, J.C.; et al. Synthesis of acyclo-C-nucleosides in the imidazo[1,2-a]pyridine and pyrimidine series as antiviral agents. J. Med. Chem. 1996, 39, 2856–2859. [Google Scholar] [CrossRef] [PubMed]
- Gueiffier, A.; Blache, Y.; Chapat, J.P.; Elhakmaoui, A.; Essassi, E.M.; Andrei, G.; Snoeck, R.; De Clercq, E.; Chavignon, O.; Teulade, J.C.; et al. Synthesis and antiviral activity of 2 and 3-substituted imidazo[1,2-a]pyrimidine. Nucleosides Nucleotides Nucleic Acids 1995, 14, 551–554. [Google Scholar] [CrossRef]
- Moog, C.; Wick, A.; Le Ber, P.; Kirn, A.; Aubertin, A.M. Bicyclic imidazo derivatives, a new class of highly selective inhibitors for the human immunodeficiency virus type 1. Antivir. Res. 1994, 24, 275–288. [Google Scholar] [CrossRef]
- Chen, X.; Xu, W.; Wang, K.; Mo, M.; Zhang, W.; Du, L.; Yuan, X.; Xu, Y.; Wang, Y.; Shen, J. Discovery of a Novel Series of Imidazo[1,2-a]pyrimidine derivatives as potent and orally bioavailable lipoprotein-associated phospholipase A2 inhibitors. J. Med. Chem. 2015, 58, 8529–8541. [Google Scholar] [CrossRef] [PubMed]
- Kishbaugh, T.L. Pyridines and Imidazopyridines with Medicinal Significance. Curr. Top. Med. Chem. 2016, 16, 3274–3302. [Google Scholar] [CrossRef] [PubMed]
- Cesur, Z.; Cesur, N.; Birteksöz, S.; Otük, G. Synthesis and biological evaluation of some new imidazo[1,2-a]pyridines. Acta Chim. Slov. 2010, 57, 355–362. [Google Scholar] [PubMed]
- Kaplancikli, Z.A.; Turan-Zitouni, G.; Ozdemir, A.; Revial, G. Synthesis and anticandidal activity of some imidazopyridine derivates. J. Enzyme Inhib. Med. Chem. 2008, 23, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M.; Bernal-Martínez, L.; Buitrago, M.J.; Castelli, M.V.; Gomez-Lopez, A.; Zaragoza, O.; Rodriguez-Tudela, J.L. Update on the epidemiology and diagnosis of invasive fungal infection. Int. J. Antimicrob. Agents 2008, 32 (Suppl. 2), 143–147. [Google Scholar] [CrossRef]
- Olaechea, P.M.; Palomar, M.; León-Gil, C.; Álvarez-Lerma, F.; Jordá, R.; Nolla-Salas, J.; León-Regidor, M.A.; EPCAN Study Group. Economic impact of Candida colonization and Candida infection in the critically ill patient. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.V.; van de Veerdonk, F.L.; Lionakis, M.S. Understanding the role of host immune responses in invasive candidiasis. Intensive Care Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabelo, V.W.; Santos, T.F.; Terra, L.; Santana, M.V.; Castro, H.C.; Rodrigues, C.R.; Abreu, P.A. Targeting CYP51 for drug design by the contributions of molecular modeling. Fundam. Clin. Pharmacol. 2017, 31, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, Z.; Khabnadideh, S.; Zomorodian, K.; Pakshir, K.; Kashi, G.; Sanagoei, N.; Gholami, S. Design, synthesis and antifungal activity of some new imidazole and triazole derivatives. Arch. Pharm. (Weinh.) 2011, 344, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, S.S.; Souza, A.C.R.; Chowdhary, A.; Meis, J.F.; Colombo, A.L. Epidemiology and molecular mechanisms of antifungal resistance in Candida and Aspergillus. Mycoses 2016, 59, 198–219. [Google Scholar] [CrossRef] [PubMed]
- Masiá-Canuto, M.; Gutiérrez-Rodero, F. Antifungal drug resistance to azoles and polyenes. Lancet Infect. Dis. 2002, 2, 550–563. [Google Scholar] [CrossRef]
- Rival, Y.; Grassy, G.; Taudou, A.; Ecalle, R. Antifungal activity in vitro of some imidazo[1,2-a]pyrimidine derivatives. Eur. J. Med. Chem. 1991, 26, 13–18. [Google Scholar] [CrossRef]
- Rival, Y.; Taudou, A.; Ecalle, R. Synthesis and antifungal activity evaluation of 3-hydroxyimidazo [1,2-a] pyridine and 3-hydroxyimidazo [1,2-a] pyrimidine derivatives. Farmaco 1993, 48, 857–869. [Google Scholar]
- El Kazzouli, S.; Berteina-Raboin, S.; Mouaddib, A.; Guillaumet, G. Solid-phase synthesis of imidazo[1,2-a]pyridines and imidazo[1,2-a]pyrimidines. Tetrahedron Lett. 2003, 44, 6265–6267. [Google Scholar] [CrossRef]
- Cosimelli, B.; Laneri, S.; Ostacolo, C.; Sacchi, A.; Severi, E.; Porcù, E.; Rampazzo, E.; Moro, E.; Basso, G.; Viola, G. Synthesis and biological evaluation of imidazo[1,2-a]pyrimidines and imidazo[1,2-a]pyridines as new inhibitors of the Wnt/β-catenin signaling. Eur. J. Med. Chem. 2014, 83, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Ermolat’ev, D.S.; Giménez, V.N.; Babaev, E.V.; Van der Eycken, E. Efficient Pd(0)-mediated microwave-assisted arylation of 2-substituted imidazo[1,2-a]pyrimidines. J. Comb. Chem. 2006, 8, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Olvera, S.; Salgado-Zamora, H.; Velázquez-Ponce, M.; Campos-Aldrete, E.; Reyes-Arellano, A.; Pérez-González, C. Fluorescent property of 3-hydroxymethyl imidazo[1,2-a]pyridine and pyrimidine derivatives. Chem. Cent. J. 2012, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Podergajs, S.; Stanovnik, B.; Tisler, M. A New Approach for the Synthesis of Fused Imidazoles: The synthesis of 3-acyl-substituted imidazo[1,2-x]azines. Synthesis 1984, 3, 263–265. [Google Scholar] [CrossRef]
- Shaaban, M.R. Facile Access to Novel 3-Acylimidazo[1,2-a]pyrimidines under Microwave Irradiation. Heterocycles 2013, 87, 1775–1783. [Google Scholar] [CrossRef]
- Gómez, O.; Salgado-Zamora, H.; Reyes, A.; Campos, M.E. A revised approach to the synthesis of 3-acyl imidazo[l,2-a]pyridines. Heterocycl. Commun. 2010, 16, 99–104. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Fitzjohn, P.W.; Bates, P.A. Comparative modelling: An essential methodology for protein structure prediction in the post-genomic era. Appl. Bioinform. 2002, 1, 177–190. [Google Scholar]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- Yates, C.M.; Garvey, E.P.; Shaver, S.R.; Schotzinger, R.J.; Hoekstra, W.J. Design and optimization of highly-selective, broad spectrum fungal CYP51 inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 3243–3248. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, W.J.; Garvey, E.P.; Moore, W.R.; Rafferty, S.W.; Yates, C.M.; Schotzinger, R.J. Design and optimization of highly-selective fungal CYP51 inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 3455–3458. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Cui, Y.L.; Wu, R.L. Molecular dynamic modeling of CYP51B in complex with azole inhibitors. J. Biomol. Struct. Dyn. 2017, 28, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M.; Lee-Yang, W.; Ciblak, M.A.; Arthington-Skaggs, B.A.; Mellado, E.; Warnock, D.W.; Rodríguez-Tudela, J.L. Comparative evaluation of NCCLS M27-A and EUCAST broth microdilution procedures for antifungal susceptibility testing of Candida species. Antimicrob. Agents Chemother. 2002, 46, 3644–4647. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.; Singh, G.; Sanz, D.; Claramunt, R.M.; Torralba, M.C.; Torres, M.R. NBS mediated one-pot regioselective synthesis of 2,3-disubstitutedimidazo[1,2-a]pyridines and their unambiguous characterization through 2D NMR and X-ray crystallography. Tetrahedron 2016, 72, 3832–3838. [Google Scholar] [CrossRef]
- Horowitz, S.; Trievel, R.C. Carbon-Oxygen Hydrogen Bonding in Biological Structure and Function. J. Biol. Chem. 2012, 287, 41576–41582. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lee, M.K.; Jefcoate, C.; Kim, S.C.; Chen, F.; Yu, J.H. Fungal cytochrome p450 monooxygenases: Their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol. Evol. 2014, 6, 1620–1634. [Google Scholar] [CrossRef] [PubMed]
- Hovmöller, S.; Zhou, T.; Ohlson, T. Conformations of amino acids in proteins. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Zhang, W.; Zhou, Y.; Zhang, M.; Zhu, J.; Song, Y.; Lü, J. A three-dimensional model of lanosterol 14alpha-demethylase of Candida albicans and its interaction with azole antifungals. J. Med. Chem. 2000, 43, 2493–2505. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Chai, X.; Hu, H.; Yan, Y.; Guan, Z.; Zou, Y.; Sun, Q.; Wu, Q. Synthesis and antifungal evaluation of novel triazole derivatives as inhibitors of cytochrome P450 14alpha-demethylase. Eur. J. Med. Chem. 2010, 45, 4435–4445. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ni, T.; Chai, X.; Wang, T.; Wang, H.; Chen, J.; Jin, Y.; Zhang, D.; Yu, S.; Jiang, Y. Molecular docking, design, synthesis and antifungal activity study of novel triazole derivatives. Eur. J. Med. Chem. 2018, 143, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- Salazar, S.B.; Wang, C.; Munsterkotter, M.; Okamoto, M.; Takahashi-Nakaguchi, A.; Chibana, H.; Lopes, M.M.; Guldener, U.; Butler, G.; Mira, N.P. Comparative Genomic and Transcriptomic Analyses Unveil Novel Features of Azole Resistance and Adaptation to the Human Host in Candida Glabrata. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Mazari, W.; Boucherit-Otmani, Z.; Boucherit, K. In Vitro Susceptibility of Amphotericin-B, Voriconazole and Caspofungin Against Candida Guilliermondii Biofilms, Isolated from Dentals Units Water Pipes, under Different Growth Phases. J. Mycol. Med. 2015, 25, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Boyken, L.; Hollis, R.J.; Kroeger, J.; Messer, S.A.; Tendolkar, S.; Diekema, D.J. Wild-Type MIC Distributions and Epidemiological Cutoff Values for Posaconazole and Voriconazole and Candida spp. as Determined by 24-Hour CLSI Broth Microdilution. J. Clin. Microbiol. 2011, 49, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Chryssanthou, E.; Cuenca-Estrella, M. Comparison of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antibiotic Susceptibility Testing Proposed Standard and the E-Test with the NCCLS Broth Microdilution Method for Voriconazole and Caspofungin Susceptibility Testing of Yeast Species. J. Clin. Microbiol. 2002, 40, 3841–3844. [Google Scholar] [PubMed]
- Chai, X.; Zhang, J.; Cao, Y.; Zou, Y.; Wu, Q.; Zhang, D.; Jiang, Y.; Sun, Q. New azoles with antifungal activity: Design, synthesis, and molecular docking. Bioorg. Med. Chem. Lett. 2011, 21, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Al-Wabli, R.I.; Al-Ghamdi, A.R.; Ghabbour, H.A.; Al-Agamy, M.H.; Monica, J.C.; Joe, I.H.; Attia, M.I. Synthesis, X-ray single crystal structure, molecular docking and DFT computations on N-[(1E)-1-(2H-1,3-benzodioxol-5-yl)-3-(1H-imidazol-1-yl)propylidene]-hydroxylamine: A new potential antifungal agent precursor. Molecules 2017, 22, 373. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2014, 47, 5–6. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment; Release 2017; Dassault Systèmes: San Diego, CA, USA, 2016.
- Laskowski, R.A. PDBsum: Summaries and analyses of PDB structures. Nucleic Acids Res. 2001, 29, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Hsin, J.; Arkhipov, A.; Yin, Y.; Stone, J.E.; Schulten, K. Using VMD: An introductory tutorial. Curr. Protoc. Bioinform. 2008. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
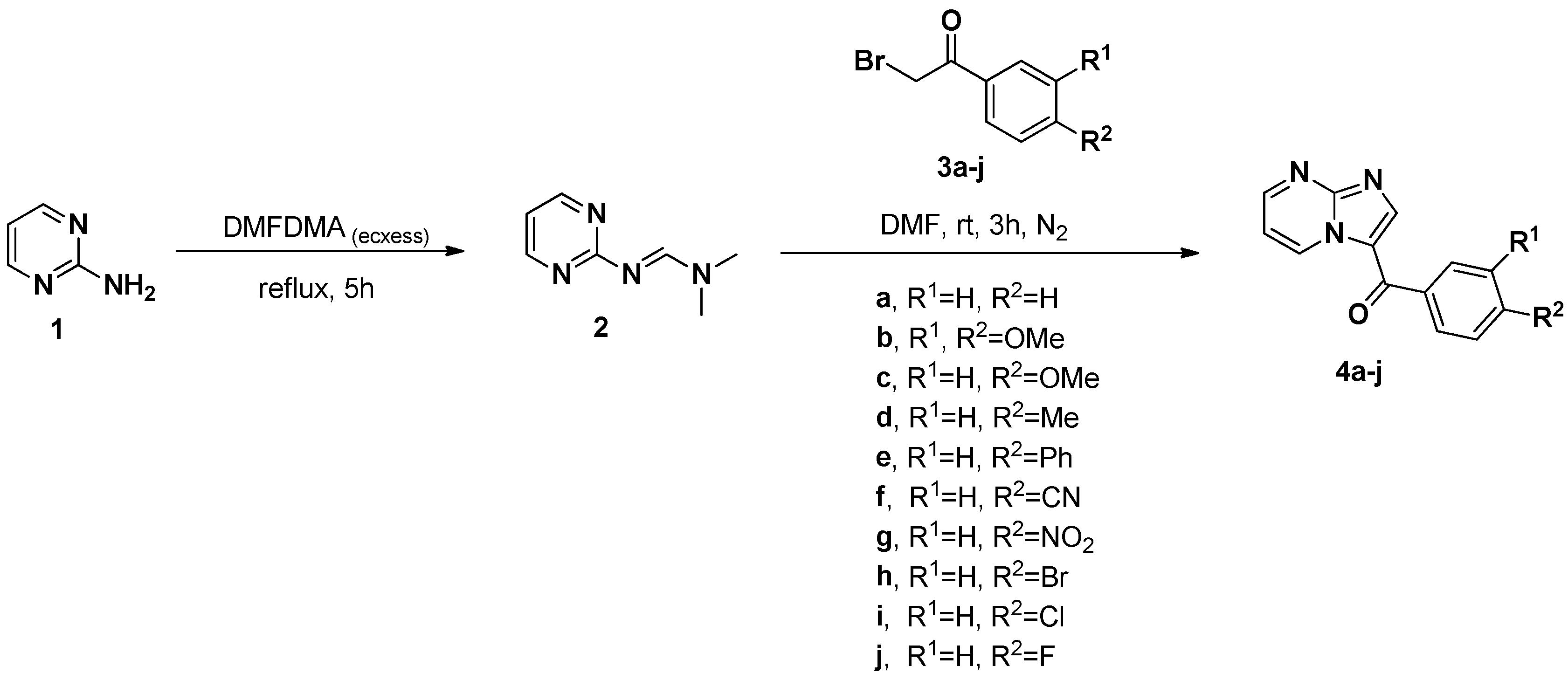


| Entry | Compound | R1 | R2 | Yield (%) |
|---|---|---|---|---|
| 1 | 4a | H | H | 77 |
| 2 | 4b | OMe | OMe | 81 |
| 3 | 4c | H | OMe | 67 |
| 4 | 4d | H | Me | 90 |
| 5 | 4e | H | Ph | 70 |
| 6 | 4f | H | CN | 84 |
| 7 | 4g | H | NO2 | 74 |
| 8 | 4h | H | Br | 98 |
| 9 | 4i | H | Cl | 72 |
| 10 | 4j | H | F | 62 |

| Compound | H-2 | H-5 | H-6 | H-7 | H-12 | H-13 | H-14 | H-15 | H-16 |
|---|---|---|---|---|---|---|---|---|---|
| 4a * | 8.41 | 10.0 | 7.23 | 8.84 | 7.90 | 7.57 | 7.65 | 7.57 | 7.90 |
| 4b * | 8.39 | 9.88 | 7.18 | 8.78 | 7.43 | 3.94 | 3.96 | 6.96 | 7.54 |
| 4c * | 8.37 | 9.90 | 7.17 | 8.78 | 7.88 | 7.00 | 3.90 | 7.00 | 7.88 |
| 4d * | 8.37 | 9.94 | 7.19 | 8.79 | 7.77 | 7.32 | 2.48 | 7.32 | 7.77 |
| 4e ** | 8.53 | 10.08 | 7.74 | 9.08 | 7.87 | 7.74 | 7.29–7.52 | 7.74 | 7.87 |
| 4f ** | 8.51 | 10.10 | 7.77 | 9.13 | 7.62 | 7.62 | - | 7.62 | 7.62 |
| 4g ** | 8.52 | 10.08 | 7.76 | 9.10 | 7.96 | 8.29 | - | 8.29 | 7.96 |
| 4h ** | 8.43 | 10.00 | 7.67 | 9.04 | 7.67 | 7.40 | - | 7.40 | 7.67 |
| 4i ** | 8.42 | 10.00 | 7.68 | 9.03 | 7.68 | 7.41 | - | 7.41 | 7.68 |
| 4j ** | 8.39 | 9.96 | 7.92–7.96 | 8.84 | 7.92–7.96 | 7.22–7.27 | - | 7.22–7.27 | 7.92–7.96 |
| Compound | Binding Energy (kcal/mol) inCYP51 from Candida spp. | ||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | |
| Fluconazole | −4.44 | −4.56 | −4.38 | −4.07 | −3.16 | −5.68 | −4.99 |
| Ketoconazole | 32.55 | −1.95 | −6.16 | −5.5 | 48.93 | 1.44 | 3.92 |
| 4a | −7.65 | −7.53 | −7.1 | −7.21 | −6.85 | −8.41 | −7.86 |
| 4d | −8.06 | −7.87 | −7.64 | −7.79 | −6.34 | −9.09 | −8.52 |
| 4f | −8.43 | −7.91 | −7.61 | −7.89 | −6.11 | −9.43 | −8.39 |
| 4i | −8.02 | −7.74 | −7.73 | −7.87 | −6.21 | −9.26 | −8.45 |
| 4j | −7.8 | −7.59 | −7.13 | −7.19 | −7.17 | −8.5 | −8.02 |
| Compound | MIC50 (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | |
| Fluconazole | 0.5 | 0.5 | 4 | 1 | 0.5 | 4 | 1 |
| Ketoconazole | 8 | 2 | 0.125 | 0.25 | 0.25 | 1 | 2 |
| 4a | 0.125 | 0.0625 | 0.0625 | 0.0625 | 0.125 | 0.5 | 0.5 |
| 4d | 0.125 | 0.25 | 0.0625 | 0.0625 | 0.5 | 0.5 | 0.25 |
| 4f | 0.25 | 0.0625 | 0.0312 | 0.0625 | 0.125 | 0.25 | 0.0625 |
| 4i | 0.25 | 0.5 | 0.0625 | 1 | 0.125 | 0.5 | 0.0312 |
| 4j | 0.0625 | 0.5 | 0.25 | 0.0312 | 0.125 | 0.5 | 0.25 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-García, O.; Andrade-Pavón, D.; Campos-Aldrete, E.; Ballinas-Indilí, R.; Méndez-Tenorio, A.; Villa-Tanaca, L.; Álvarez-Toledano, C. Synthesis, Molecular Docking, and Antimycotic Evaluation of Some 3-Acyl Imidazo[1,2-a]pyrimidines. Molecules 2018, 23, 599. https://doi.org/10.3390/molecules23030599
Gómez-García O, Andrade-Pavón D, Campos-Aldrete E, Ballinas-Indilí R, Méndez-Tenorio A, Villa-Tanaca L, Álvarez-Toledano C. Synthesis, Molecular Docking, and Antimycotic Evaluation of Some 3-Acyl Imidazo[1,2-a]pyrimidines. Molecules. 2018; 23(3):599. https://doi.org/10.3390/molecules23030599
Chicago/Turabian StyleGómez-García, Omar, Dulce Andrade-Pavón, Elena Campos-Aldrete, Ricardo Ballinas-Indilí, Alfonso Méndez-Tenorio, Lourdes Villa-Tanaca, and Cecilio Álvarez-Toledano. 2018. "Synthesis, Molecular Docking, and Antimycotic Evaluation of Some 3-Acyl Imidazo[1,2-a]pyrimidines" Molecules 23, no. 3: 599. https://doi.org/10.3390/molecules23030599