Computational Insight into the Effect of Natural Compounds on the Destabilization of Preformed Amyloid-β(1–40) Fibrils
Abstract
:1. Introduction
2. Methods
2.1. Molecular Dynamics Simulations
2.2. Ligand Binding Sites
2.3. Ligand Binding Energy
2.4. Aβ(1–40) Oligomer Double-Layered Structures
3. Results and Discussion
3.1. Putative Binding Sites and Binding Free Energies
- β-1 β-sheet corresponding to the amino-acid sequence: 16KLVFFAEDV24,
- β-2 β-sheet corresponding to the amino-acid sequence: 31IIGLMVG37,
- Elbow connecting the two β-sheets with the corresponding amino-acid sequence: 22EDVGSN27,
- top of the protofibril, over the two β-sheets of the terminal Aβ(1–40) monomer (“Over”),
- disordered tails located at the N-terminal,
- end of the β-2 β-sheet, on the C-terminal (entry of the cleft).
- -
- CUR-ke, the predominant form in aqueous solution on the basis of the recent results obtained by Manolova et al. [54], shows a strong propensity to dock at the β-2 site and realizes at this site strong interactions (ΔGbind > −20 kcal/mol) with the fAβ(1–40) fibril. However, moderate to strong (−10 < ΔGbind > −20 kcal/mol) free energies of binding are found for all the binding sites, with the exception of the N-terminal (N-ter) one.
- -
- DOPA shows a preference for docking at the β-2 and N-ter sites. However, by considering the free energy of binding, it does not show selectivity among the six sites studied, realizing moderate to weak interactions (Gbind < −10 kcal/mol) with all of them.
- -
- EGCG preferentially targets the N-ter and β-2 sites and secondarily, the Elbow and β-1 sites. However, this ligand is able to realize strong binding with all six possible sites. The most stable complexes (ΔGbind > −20 kcal/mol) are obtained at the β-2 and β-1 sites. The ability of EGCG to bind to the N-terminal amino acids (residues 1–16) is confirmed by results obtained by isothermal titration calorimetry experiments [55]. Moreover, recent findings by solution NMR indicate that EGCG preferentially binds to Aβ oligomers and shields them at the β-1 and β-2 sites [56], where it remodels the oligomer surface, altering the interactions with the monomers.
- -
- QUER is found almost equally distributed between the β-1 and Over sites, with significantly lower probability for the other sites. However, it realizes moderate binding free energies (ΔGbind~−10 kcal/mol) in all sites, with the most stable complexes (ΔGbind~−20 kcal/mol) involving the β-2 and β-1 sites. These results are in agreement with the finding of a computational study recently reported by Ren et al. [13] for a structurally homologous compound, genistein. They showed that genistein prefers to bind the β-sheet grooves to interfere with their self-aggregation.
- -
- ROSM has higher probability for docking at the N-ter and β-1 sites, but realizes the most stable interactions with moderate binding free energies at the β-2 and β-1 sites. Indeed, NMR investigations suggest that a ROSM hairpin-like structure would allow the intercalation into the Aβ oligomers structure at the interprotofilament (β−β zippers) interface [57].
3.2. Influence of the Ligands on the Stability of the Aβ(1–40) Oligomer Double-Layered Structures
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; He, G. Toward a Rational Design to Regulate β-Amyloid Fibrillation for Alzheimer’s Disease Treatment. ACS Chem. Neurosci. 2018, 9, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Yair, P.; Adel, A.; Ehud, G. Inhibition of Amyloid Fibril Formation by Polyphenols: Structural Similarity and Aromatic Interactions as a Common Inhibition Mechanism. Chem. Biol. Drug Des. 2005, 67, 27–37. [Google Scholar] [CrossRef]
- Lemkul, J.A.; Bevan, D.R. The Role of Molecular Simulations in the Development of Inhibitors of Amyloid β-Peptide Aggregation for the Treatment of Alzheimer’s Disease. ACS Chem. Neurosci. 2012, 3, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.-F.; Dong, X.-Y.; He, L.; Middelberg, A.P.J.; Sun, Y. Molecular Insight into Conformational Transition of Amyloid β-Peptide 42 Inhibited by (−)-Epigallocatechin-3-gallate Probed by Molecular Simulations. J. Phys. Chem. B 2011, 115, 11879–11887. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.N.; Chiu, S.-W.; Benoit, J.; Chew, L.Y.; Mu, Y. The Effect of Curcumin on the Stability of Aβ Dimers. J. Phys. Chem. B 2012, 116, 7428–7435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, J.; Derreumaux, P.; Mu, Y. Molecular Mechanism of the Inhibition of EGCG on the Alzheimer Aβ1–42 Dimer. J. Phys. Chem. B 2013, 117, 3993–4002. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Latshaw, D.C.; Hall, C.K. Aggregation of Aβ(17–36) in the Presence of Naturally Occurring Phenolic Inhibitors Using Coarse-Grained Simulations. J. Mol. Biol. 2017, 429, 3893–3908. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, M.; Singh, S.; Pandey, V.P.; Dwivedi, U.N. Modulation in the conformational and stability attributes of the Alzheimer’s disease associated amyloid-beta mutants and their favorable stabilization by curcumin: Molecular dynamics simulation analysis. J. Biomol. Struct. Dyn. 2017, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chebaro, Y.; Jiang, P.; Zang, T.; Mu, Y.; Nguyen, P.H.; Mousseau, N.; Derreumaux, P. Structures of Aβ17–42 Trimers in Isolation and with Five Small-Molecule Drugs Using a Hierarchical Computational Procedure. J. Phys. Chem. B 2012, 116, 8412–8422. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.P.N.; Mohamed, T.; Teckwani, K.; Tin, G. Curcumin Binding to Beta Amyloid: A Computational Study. Chem. Biol. Drug Des. 2015, 86, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Liu, Y.; Zhang, Y.; Cai, Y.; Gong, X.; Chang, Y.; Xu, L.; Zheng, J. Genistein: A Dual Inhibitor of Both Amyloid β and Human Islet Amylin Peptides. ACS Chem. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, R.; Hatayama, K.; Takahashi, T.; Hayashi, T.; Sato, Y.; Sato, D.; Ohta, K.; Nakano, H.; Seki, C.; Endo, Y.; Tokuraku, K.; Uwai, K. Structure–activity relations of rosmarinic acid derivatives for the amyloid β aggregation inhibition and antioxidant properties. Eur. J. Med. Chem. 2017, 138, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Battisti, A.; Piccionello, A.P.; Sgarbossa, A.; Vilasi, S.; Ricci, C.; Ghetti, F.; Spinozzi, F.; Gammazza, A.M.; Giacalone, V.; Martorana, A.; Lauria, A.; Ferrero, C.; Bulone, D.; Rosalia Mangione, M.; Biagio, P.L.S.; Grazia Ortore, M. Curcumin-like compounds designed to modify amyloid beta peptide aggregation patterns. RSC Adv. 2017, 7, 31714–31724. [Google Scholar] [CrossRef]
- Espargaró, A.; Ginex, T.; Vadell, M.D.; Busquets, M.A.; Estelrich, J.; Muñoz-Torrero, D.; Luque, F.J.; Sabate, R. Combined in Vitro Cell-Based/in Silico Screening of Naturally Occurring Flavonoids and Phenolic Compounds as Potential Anti-Alzheimer Drugs. J. Nat. Prod. 2017, 80, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Li, M.S. Curcumin Binds to Aβ1–40 Peptides and Fibrils Stronger Than Ibuprofen and Naproxen. J. Phys. Chem. B 2012, 116, 10165–10175. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Fang, S.-T.; Huang, S.-H.; Chou, C.-L.; Huy, P.D.Q.; Li, M.S.; Chen, Y.-C. Anti-arrhythmic Medication Propafenone a Potential Drug for Alzheimer’s Disease Inhibiting Aggregation of Aβ: In Silico and in Vitro Studies. J. Chem. Inf. Model. 2016, 56, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Kawai, R.; Araki, M.; Yoshimura, M.; Kamiya, N.; Ono, M.; Saji, H.; Okuno, Y. Core Binding Site of a Thioflavin-T-Derived Imaging Probe on Amyloid β Fibrils Predicted by Computational Methods. ACS Chem. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Murugan, N.A.; Halldin, C.; Nordberg, A.; Långström, B.; Ågren, H. The Culprit Is in the Cave: The Core Sites Explain the Binding Profiles of Amyloid-Specific Tracers. J. Phys. Chem. Lett. 2016, 7, 3313–3321. [Google Scholar] [CrossRef] [PubMed]
- Peccati, F.; Pantaleone, S.; Riffet, V.; Solans-Monfort, X.; Contreras-García, J.; Guallar, V.; Sodupe, M. Binding of Thioflavin T and Related Probes to Polymorphic Models of Amyloid-β Fibrils. J. Phys. Chem. B 2017, 121, 8926–8934. [Google Scholar] [CrossRef] [PubMed]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. EBJ 2011, 40, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Petkova, A.T.; Yau, W.-M.; Tycko, R. Experimental Constraints on Quaternary Structure in Alzheimer’s β-Amyloid Fibrils†. Biochemistry (Mosc.) 2006, 45, 498–512. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucl. Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.-Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2006, 15, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; Hess, B.; Lindahl, E. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinforma. Oxf. Engl. 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Koziara, K.B.; Stroet, M.; Malde, A.K.; Mark, A.E. Testing and validation of the Automated Topology Builder (ATB) version 2.0: Prediction of hydration free enthalpies. J. Comput. Aided Mol. Des. 2014, 28, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput. 2011, 7, 4026–4037. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces: Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry Held in Jerusalem, Israel, April 13–16, 1981; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. ISBN 978-94-015-7658-1. [Google Scholar]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Patriksson, A.; Spoel, D. van der A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys. 2008, 10, 2073–2077. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Hung, H.M.; Tran, K.N.; Nguyen, M.T. Replica exchange molecular dynamics study of the amyloid beta (11–40) trimer penetrating a membrane. RSC Adv. 2017, 7, 7346–7357. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Shrake, A.; Rupley, J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973, 79, 351–371. [Google Scholar] [CrossRef]
- Sitkoff, D.; Sharp, K.A.; Honig, B. Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models. J. Phys. Chem. 1994, 98, 1978–1988. [Google Scholar] [CrossRef]
- Sharp, K.A.; Honig, B. Electrostatic interactions in macromolecules: Theory and applications. Annu. Rev. Biophys. Biophys. Chem. 1990, 19, 301–332. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.; Hünenberger, P.H.; McCammon, J.A. Absolute Single-Molecule Entropies from Quasi-Harmonic Analysis of Microsecond Molecular Dynamics: Correction Terms and Convergence Properties. J. Chem. Theory Comput. 2009, 5, 3150–3160. [Google Scholar] [CrossRef] [PubMed]
- Tycko, R. Molecular structure of amyloid fibrils: Insights from solid-state NMR. Q. Rev. Biophys. 2006, 39, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Tycko, R. Amyloid Polymorphism: Structural Basis and Neurobiological Relevance. Neuron 2015, 86, 632–645. [Google Scholar] [CrossRef] [PubMed]
- Trzesniak, D.; Kunz, A.P.; van Gunsteren, W.F. A Comparison of Methods to Compute the Potential of Mean Force. ChemPhysChem 2006, 8, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Roux, B. The calculation of the potential of mean force using computer simulations. Comput. Phys. Commun. 1995, 91, 275–282. [Google Scholar] [CrossRef]
- Ono, K.; Hasegawa, K.; Naiki, H.; Yamada, M. Curcumin has potent anti-amyloidogenic effects for Alzheimer’s β-amyloid fibrils in vitro. J. Neurosci. Res. 2004, 75, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lim, G.P.; Begum, A.N.; Ubeda, O.J.; Simmons, M.R.; Ambegaokar, S.S.; Chen, P.P.; Kayed, R.; Glabe, C.G.; Frautschy, S.A.; et al. Curcumin inhibits formation of amyloid beta oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 2005, 280, 5892–5901. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Yoshiike, Y.; Takashima, A.; Hasegawa, K.; Naiki, H.; Yamada, M. Potent anti-amyloidogenic and fibril-destabilizing effects of polyphenols in vitro: Implications for the prevention and therapeutics of Alzheimer’s disease. J. Neurochem. 2003, 87, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Bieschke, J.; Russ, J.; Friedrich, R.P.; Ehrnhoefer, D.E.; Wobst, H.; Neugebauer, K.; Wanker, E.E. EGCG remodels mature α-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. 2010, 107, 7710–7715. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Hasegawa, K.; Naiki, H.; Yamada, M. Anti-Parkinsonian agents have anti-amyloidogenic activity for Alzheimer’s beta-amyloid fibrils in vitro. Neurochem. Int. 2006, 48, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Kundaikar, H.S.; Degani, M.S. Insights into the Interaction Mechanism of Ligands with Aβ42 Based on Molecular Dynamics Simulations and Mechanics: Implications of Role of Common Binding Site in Drug Design for Alzheimer’s Disease. Chem. Biol. Drug Des. 2015, 86, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Fukuchi, M.; Yatagawa, T.; Tada, M.; Takeda, K.; Irie, K.; Akagi, K.; Monobe, Y.; Imazawa, T.; Takegoshi, K. Solid-state NMR analysis of interaction sites of curcumin and 42-residue amyloid β-protein fibrils. Bioorg. Med. Chem. 2011, 19, 5967–5974. [Google Scholar] [CrossRef] [PubMed]
- Manolova, Y.; Deneva, V.; Antonov, L.; Drakalska, E.; Momekova, D.; Lambov, N. The effect of the water on the curcumin tautomerism: A quantitative approach. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2014, 132, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-H.; Dong, X.-Y.; Sun, Y. Thermodynamic Analysis of the Molecular Interactions between Amyloid β-Protein Fragments and (−)-Epigallocatechin-3-gallate. J. Phys. Chem. B 2012, 116, 5803–5809. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Melacini, G. A solution NMR toolset to probe the molecular mechanisms of amyloid inhibitors. Chem. Commun. 2018. [Google Scholar] [CrossRef] [PubMed]
- Airoldi, C.; Sironi, E.; Dias, C.; Marcelo, F.; Martins, A.; Rauter, A.P.; Nicotra, F.; Jimenez-Barbero, J. Natural Compounds against Alzheimer’s Disease: Molecular Recognition of Aβ1–42 Peptide by Salvia sclareoides Extract and its Major Component, Rosmarinic Acid, as Investigated by NMR. Chem. Asian J. 2013, 8, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Gonnelli, L.; Luchinat, C.; Mao, J.; Nesi, A. A New Structural Model of Aβ40 Fibrils. J. Am. Chem. Soc. 2011, 133, 16013–16022. [Google Scholar] [CrossRef] [PubMed]
- Korn, A.; McLennan, S.; Adler, J.; Krueger, M.; Surendran, D.; Maiti, S.; Huster, D. Amyloid β (1-40) Toxicity Depends on the Molecular Contact between Phenylalanine 19 and Leucine 34. ACS Chem. Neurosci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Jang, H.; Ma, B.; Tsai, C.-J.; Nussinov, R. Modeling the Alzheimer Aβ17-42 Fibril Architecture: Tight Intermolecular Sheet-Sheet Association and Intramolecular Hydrated Cavities. Biophys. J. 2007, 93, 3046–3057. [Google Scholar] [CrossRef] [PubMed]
- Berhanu, W.M.; Hansmann, U.H.E. Structure and Dynamics of Amyloid-β Segmental Polymorphisms. PLoS ONE 2012, 7, e41479. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available. |





| Compound | Acronym | Structure | Aβ(1–40) Formation (EC50) μM | Aβ(1–40) Extension (EC50) μM | Aβ(1–40) Destabilization (EC50) μM |
|---|---|---|---|---|---|
| Curcumin diketo form | CUR-di |  | 0.19 [47] | 0.19 [47] | 0.42 [47] |
| Curcumin ketoenol form | CUR-ke | 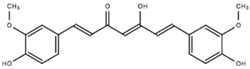 | 0.81 [48] | 0.19 [47] | 1.00 [48] |
| Dopamine | DOPA | 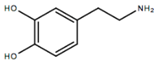 | 0.01 [51] | 0.03 [51] | 0.21 [51] |
| (-)-Epigallocatechin-3-gallate | EGCG |  | 0.18 [4] | - | 15 * [50] |
| Quercetin | QUER |  | 0.24 [49] | 0.25 [49] | 2.1 [49] |
| Rosmarinic acid | ROSM | 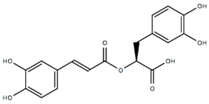 | 0.29 [47] | 0.26 [47] | 0.83 [47] |
| Force Direction | Lateral (x-axis) | Vertical (y-axis) | Outward (z-axis) | |||
|---|---|---|---|---|---|---|
| Ligand/binding site | β-1 | β-2 | β-1 | β-2 | β-1 | β-2 |
| Control | 2743 ± 115 | 2772 ± 140 | 3520 ± 200 | 2013 ± 30 | 3413 ± 250 | 2387 ± 330 |
| CUR-di | 2573 ± 40 | 1913 ± 110 | 1570 ± 70 | 1843 ± 35 | 2810 ± 10 | 2107 ± 140 |
| CUR-ke | 2600 ± 100 | 2167 ± 280 | 1653 ± 60 | 1733 ± 150 | 2760 ± 70 | 2633 ± 250 |
| DOPA | 2356 ± 95 | 2180 ± 190 | 1663 ± 55 | 1927 ± 420 | 2150 ± 95 | 2540 ± 90 |
| EGCG | 2968 ± 93 | 2407 ± 75 | 1967 ± 25 | 1610 ± 115 | 2570 ± 30 | 2533 ± 60 |
| QUER | 2493 ± 90 | 2043 ± 155 | 1726 ± 75 | 1720 ± 30 | 2553 ± 120 | 2650 ± 100 |
| ROSM | 2888 ± 173 | 1677 ± 55 | 2053 ± 40 | 1367 ± 15 | 2767 ± 70 | 2310 ± 105 |
| β-1 | β-2 | |
|---|---|---|
| Control | 14.3 ± 0.3 | 9.1 ± 0.3 |
| CUR-di | 15.7 ± 0.4 | 13.4 ± 0.3 |
| CUR-ke | 15.4 ± 0.3 | 13.6 ± 0.4 |
| DOPA | 16.5 ± 0.5 | 14.1 ± 0.4 |
| EGCG | 16.6 ± 0.5 | 13.0 ± 0.4 |
| QUER | 16.3 ± 0.4 | 12.7 ± 0.4 |
| ROSM | 15.8 ± 0.3 | 14.1 ± 0.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavanti, F.; Pedone, A.; Menziani, M.C. Computational Insight into the Effect of Natural Compounds on the Destabilization of Preformed Amyloid-β(1–40) Fibrils. Molecules 2018, 23, 1320. https://doi.org/10.3390/molecules23061320
Tavanti F, Pedone A, Menziani MC. Computational Insight into the Effect of Natural Compounds on the Destabilization of Preformed Amyloid-β(1–40) Fibrils. Molecules. 2018; 23(6):1320. https://doi.org/10.3390/molecules23061320
Chicago/Turabian StyleTavanti, Francesco, Alfonso Pedone, and Maria Cristina Menziani. 2018. "Computational Insight into the Effect of Natural Compounds on the Destabilization of Preformed Amyloid-β(1–40) Fibrils" Molecules 23, no. 6: 1320. https://doi.org/10.3390/molecules23061320







