An In Vitro Estimation of the Cytotoxicity and Genotoxicity of Root Extract from Leonurus sibiricus L. Overexpressing AtPAP1 against Different Cancer Cell Lines
Abstract
:1. Introduction
2. Results
2.1. Determination of Phenolic Acids Content in AtPAP1 and TR Root Extracts by HPLC Analysis
2.2. Assessment of Cytotoxic Activity
2.3. Effect of TR and AtPAP1 TR Root Extracts from L. sibiricus on Mitochondrial Membrane Potential Loss (ΛΨm) in K-562, CCRF-CEM and A549 Cells
2.4. Mitochondrial Copy Number
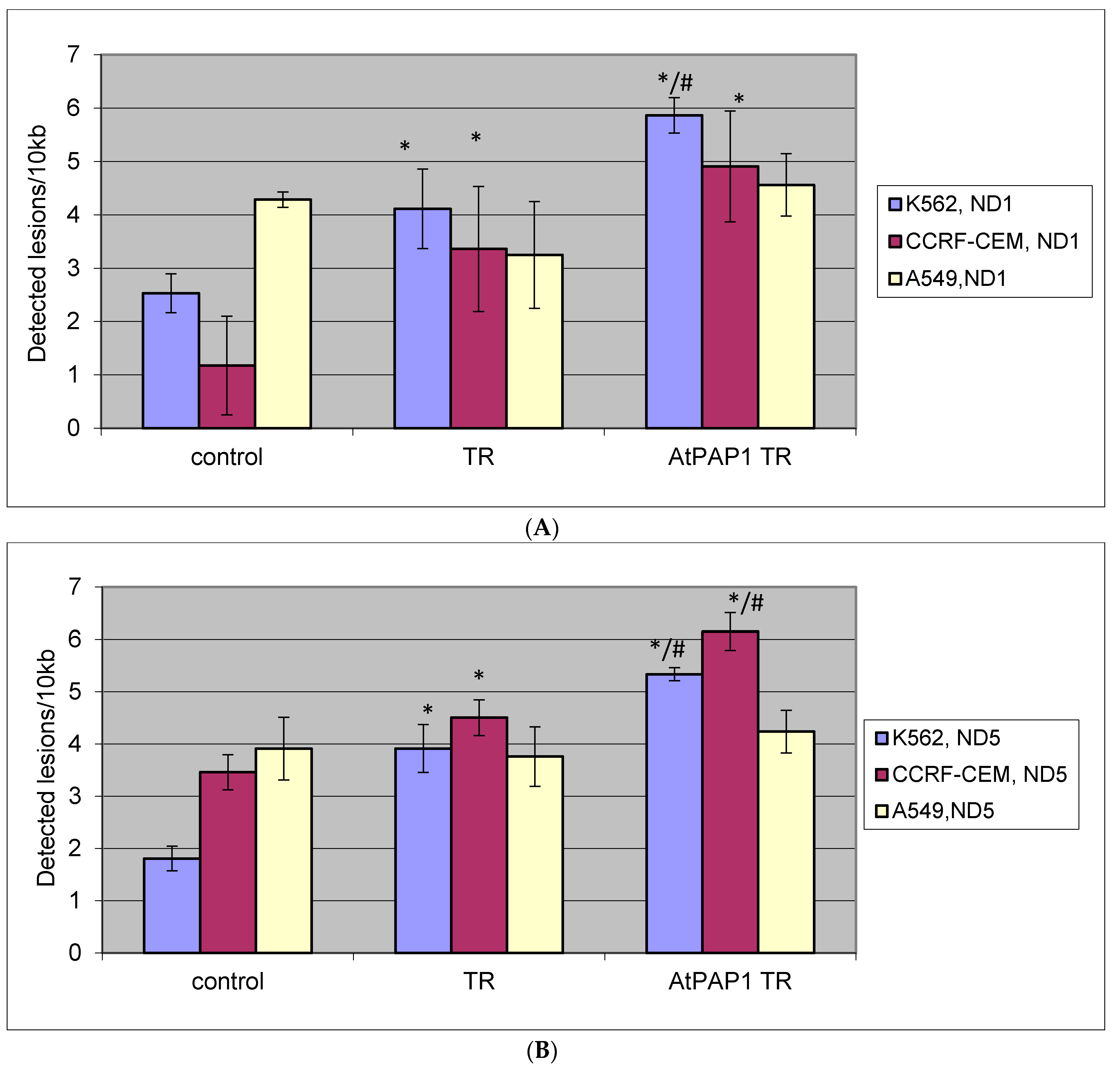
2.5. Quantification of Mitochondrial DNA (mtDNA) Damage
2.6. Quantification of Nuclear DNA (nDNA) Damage
3. Discussion
4. Materials and Methods
4.1. Establishment and Confirmation of L. sibiricus Transgenic Root Culture with Transcriptional Factor AtPAP1
4.2. Plant Material and Extract Preparation
4.3. HPLC Analysis for Determination of Phenolic Acids
4.4. Cell Cultures
4.5. Cell Viability
4.6. Mitochondrial Membrane Potential (MMP)
4.7. DNA Extraction from Cell Culture
4.8. mtDNA Copy Number Quantification
4.9. Determination of Mitochondrial and Nuclear DNA Damage
5. Statistical Analysis
6. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ochwang′I, D.O.; Kimwele, C.N.; Oduma, J.A.; Gathumbi, P.K.; Mbaria, J.M.; Kiama, S.G. Medicinal plants used in treatment and management of cancer in Kakamega County Kenya. J. Ethnopharmacol. 2014, 151, 1040–1055. [Google Scholar] [CrossRef] [PubMed]
- Greenwell, M.; Rahman, P.K.S.M. Medicinal Plants: Their Use in Anticancer Treatment. Int. J. Pharm. Sci. Res. 2015, 6, 4103–4112. [Google Scholar] [CrossRef]
- Desai, A.G.; Qazi, G.N.; Ganju, R.K.; El-Tamer, M.; Singh, J.; Saxena, A.K.; Bedi, Y.S.; Taneja, S.C.; Bhat, H.K. Medicinal Plants and Cancer Chemoprevention. Curr. Drug Metab. 2008, 9, 581–591. [Google Scholar] [CrossRef]
- Schnekenburger, M.; Dicato, M.; Diederich, M. Plant-derived epigenetic modulators for cancer treatment and prevention. Biotechnol. Adv. 2014, 32, 1123–1132. [Google Scholar] [CrossRef]
- Butler, M.S. The role of natural product chemistry in drug discovery. J. Nat. Prod. 2004, 67, 2141–2153. [Google Scholar] [CrossRef]
- Cragg, G.M.; Grothaus, P.G.; Newman, D.J. Impact of Natural products on developing new anti-cancer agents. Chem. Rev. 2009, 109, 3012–3043. [Google Scholar] [CrossRef]
- Bonam, S.R.; Wu, Y.S.; Tunki, L.; Chellian, R.; Halmuthur, M.S.K.; Muller, S.; Pandy, V. What has come out from phytomedicines and herbal edibles for the treatment of cancer? ChemMedChem 2018. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, M.; Kapulnik, Y.; Koltai, H. Plant derived substances with anti-cancer activity: From folklore to practice. Front. Plant Sci. 2015, 6, 799. [Google Scholar]
- Seca, A.M.L.; Pinto, D.C.G.A. Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef]
- Sitarek, P.; Rijo, P.; Garcia, C.; Skała, E.; Kalemba, D.; Białas, A.J.; Szemraj, J.; Pytel, D.; Toma, M.; Wysokińska, H.; et al. Antibacterial, Anti-Inflammatory, Antioxidant, and Antiproliferative Properties of Essential Oils from Hairy and Normal Roots of Leonurus sibiricus L. and Their Chemical Composition. Oxid. Med. Cell Longev. 2017, 7384061. [Google Scholar] [CrossRef] [PubMed]
- Sitarek, P.; Skała, E.; Toma, M.; Wielanek, M.; Szemraj, J.; Skorski, T.; Białas, A.J.; Sakowicz, T.; Kowalczyk, T.; Radek, M.; et al. Transformed Root Extract of Leonurus sibiricus Induces Apoptosis through Intrinsic and Extrinsic Pathways in Various Grades of Human Glioma Cells. Pathol. Oncol. Res. 2017, 23, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Sitarek, P.; Skała, E.; Wysokińska, H.; Wielanek, M.; Szemraj, J.; Toma, M.; Śliwiński, T. The Effect of Leonurus sibiricus Plant Extracts on Stimulating Repair and Protective Activity against Oxidative DNA Damage in CHO Cells and Content of Phenolic Compounds. Oxid Med. Cell Longev. 2016, 2016, 5738193. [Google Scholar] [CrossRef] [PubMed]
- Sitarek, P.; Kowalczyk, T.; Rijo, P.; Białas, A.J.; Wielanek, M.; Wysokińska, H.; Garcia, C.; Toma, M.; Śliwiński, T.; Skała, E. Over-Expression of AtPAP1 Transcriptional Factor Enhances Phenolic Acid Production in Transgenic Roots of Leonurus sibiricus L. and Their Biological Activities. Mol. Biotechnol. 2018, 60, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.S.; Cercato, L.M.; de Santana Souza, M.T.; Melo, A.J.O.; Lima, B.D.S.; Duarte, M.C.; Araujo, A.A.S.; de Oliveira E Silva, A.M.; Camargo, E.A. The ethanol extract of Leonurus sibiricus L. induces antioxidant, antinociceptive and topical anti-inflammatory effects. J. Ethnopharmacol. 2017, 206, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Sitarek, P.; Kowalczyk, T.; Picot, L.; Michalska-Hejduk, D.; Bijak, M.; Białas, A.J.; Wielanek, M.; Śliwiński, T.; Skała, E. Growth of Leonurus sibiricus L. roots with over-expression of AtPAP1 transcriptional factor in closed bioreactor, production of bioactive phenolic compounds and evaluation of their biological activity. Ind. Crops Prod. 2018, 122, 732–739. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Sitarek, P.; Kowalczyk, T.; Santangelo, S.; Białas, A.J.; Toma, M.; Wieczfinska, J.; Śliwiński, T.; Skała, E. The Extract of Leonurus sibiricus Transgenic Roots with AtPAP1 Transcriptional Factor Induces Apoptosis via DNA Damage and Down Regulation of Selected Epigenetic Factors in Human Cancer Cells. Neurochem. Res. 2018, 43, 1363–1370. [Google Scholar] [CrossRef]
- Pucci, B.; Kasten, M.; Giordano, A. Cell Cycle and Apoptosis. Neoplasia 2000, 2, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Rothfuss, O.; Gasser, T.; Patenge, N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 2010, 38, e24. [Google Scholar] [CrossRef] [PubMed]
- Afsar, T.; Trembley, J.H.; Salomon, C.E.; Razak, S.; Khan, M.R.; Ahmed, K. Growth inhibition and apoptosis in cancer cells induced by polyphenolic compounds of Acacia hydaspica: Involvement of multiple signal transduction pathways. Sci. Rep. 2016, 6, 23077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, A.; Ghorban, A. Cancer therapy with phytochemicals: Evidence from clinical studies. Avicenna J. Phytomed. 2015, 5, 84–97. [Google Scholar] [PubMed]
- Verpoorte, R.; Contin, A.; Memelink. Biotechnology for the production of plant secondary metabolites. J. Phytochem. Rev. 2002, 1, 13. [Google Scholar] [CrossRef]
- Ochoa-Villarreal, M.; Howat, S.; Hong, S.; Jang, M.O.; Young-Woo, J.; Lee, E.K.; Loake, G.J. Plant cell culture strategies for the production of natural products. BMB Rep. 2016, 49, 149–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, X.; Shen, L. Molecular targeted therapy of cancer: The progress and future prospect. Front. Lab. Med. 2017, 1, 69–75. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Chipuk, J.E.; Green, D.R. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008, 18, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Youle, R.J. The Role of Mitochondria in Apoptosis. Annu. Rev. Genet. 2009, 43, 95–118. [Google Scholar] [CrossRef] [PubMed]
- Formica, J.V.; Regelson, W. Review of the biology of quercetin and related bioflavonoids. Food Chem. Toxicol. 1995, 33, 1061–1080. [Google Scholar] [CrossRef]
- Chahar, M.K.; Sharma, N.; Dobhal, M.P.; Joshi, Y.C. Flavonoids: A versatile source of anticancer drugs. Pharmacogn. Rev. 2012, 5, 1–12. [Google Scholar]
- Plochmann, K.; Korte, G.; Koutsilieri, E. Structure-activity relationships of flavonoid-induced cytotoxicity on human leukemia cells. Arch. Biochem. Biophys. 2007, 460, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Czerwonka, A.; Kawka, K.; Cykier, K.; Lemieszek, M.K.; Rzeski, W. Evaluation of anticancer activity of water and juice extracts of young Hordeum vulgare in human cancer cell lines HT-29 and A549. Ann. Agric. Environ. Med. 2017, 24, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Ly, J.D.; Grubb, D.R.; Lawen, A. The mitochondrial membrane potential (Δψm) in apoptosis; An update. Apoptosis 2003, 8, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Yen, G.C.; Chen, C.S.; Chang, W.T.; Wu, M.F.; Cheng, F.T.; Shiau, D.K.; Hsu, C.L. Antioxidant activity and anticancer effect of ethanolic and aqueous extracts of the roots of Ficus beecheyana and their phenolic components. J. Food Drug Anal. 2018, 26, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; Liu, Z.; Fan, R.; Yin, Z.; Mi, Y.; Ren, B.; Liu, X. Athyrium multidentatum (Doll.) Ching extract induce apoptosis via mitochondrial dysfunction and oxidative stress in HepG2 cells. Sci. Rep. 2017, 7, 2275. [Google Scholar] [CrossRef] [PubMed]
- Bogenhagen, D.; Clayton, D.A. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell 1977, 11, 719–727. [Google Scholar] [CrossRef]
- Kai, Y.; Takamatsu, C.; Tokuda, K.; Okamoto, M.; Irita, K.; Takahashi, S. Rapid and random turnover of mitochondrial DNA in rat hepatocytes of primary culture. Mitochondrion 2006, 6, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Yin, P.H.; Lu, C.Y.; Chi, C.W.; Wei, Y.H. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem. J. 2000, 1, 425–432. [Google Scholar] [CrossRef]
- Chen, X.J.; Butow, R.A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005, 6, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Bijak, M.; Synowiec, E.; Sitarek, P.; Sliwiński, T.; Saluk-Bijak, J. Evaluation of the Cytotoxicity and Genotoxicity of Flavonolignans in Different Cellular Models. Nutrients 2017, 14, 1356. [Google Scholar] [CrossRef] [PubMed]
- Mizumachi, T.; Suzuki, S.; Naito, A.; Carcel-Trullols, J.; Evans, T.T.; Spring, P.M. Increased mitochondrial DNA induces acquired docetaxel resistance in head and neck cancer cells. Oncogene 2008, 7, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Basri, D.F.; Alamin, Z.A.Z.; Chan, K.M. Assessment of cytotoxicity and genotoxicity of stem bark extracts from Canarium odontophyllum Miq. (dabai) against HCT116 human colorectal cancer cell line. BMC Complement. Altern. Med. 2016, 16, 36. [Google Scholar]
- Arı, F.; Çelikler, S.; Karakaş, D.; Cevatemre, B.; Fırat, M.; Ulukaya, E. Total Phenolic Content, Antioxidant and Cyto-/Genotoxic Activities of Pelargonium quercetorum Agnew in Human Breast Cancer Cells. J. Clin. Exp. Investig. 2017, 8, 22–30. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |





| Phenolic Compounds | TR Extract (µg/g DW) | AtPAP1 TR Transgenic Extract from Bioreactor (µg/g DW) |
|---|---|---|
| Neochlorogenic acid | 8 ± 0.4 a | 7 ± 1 a |
| Chlorogenic acid | 4104 ± 8.7 a | 21,520 ± 1160 c |
| Caffeic acid | 4176 ± 9.0 a | 14,500 ± 600 d |
| p-coumaric acid | 30 ± 0.1 a | 20 ± 4 a |
| Ferulic acid | 660 ± 27.1 a | 1870 ± 160 c |
| Genome | Target Gene | Forward Primer Sequences (5′→3′) | Reverse Primer Sequence (5′→3′) | Amplicon Length (bp) |
|---|---|---|---|---|
| Mitochondrial | ND1 (mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 1) | Long fragment: ATGGCCAACCTCCTACTCCT | Long fragment: GATGAGTGTGCCTGCAAAGA | 1214 |
| Small fragment: CCTAAAACCCGCCACATCTA | Small fragment: GCCTAGGTTGAGGTTGACCA | 124 | ||
| ND5 (mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5) | Long fragment: TCCAACTCATGAGACCCACA | Long fragment: AGGTGATGATGGAGGTGGAG | 1156 | |
| Small fragment: AGGCGCTATCACCACTCTGT | Small fragment: TTGGTTGATGCCGATTGTAA | 124 | ||
| Nuclear | TP53 (tumor protein p53) | Long fragment: GGGTGTAGATGATGGGGATG | Long fragment: AACTGCGGAATGAAACAACC | 1172 |
| Small fragment: AAGCTGCTAAGGTCCCACAA | Small fragment: GGAAAGATCGCTCCAGGAA | 56 | ||
| HPRT1 (hypoxanthine phosphoribosyltransferase 1) | Long fragment: AGGGCAAAGGATGTGTTACG | Long fragment: AGTGGTTTCTGGTGCGACTT | 1018 | |
| Small fragment: TGCTGACCTGCTGGATTACA | Small fragment: TCTACAGTCATAGGAATGGATCTATCA | 69 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sitarek, P.; Synowiec, E.; Kowalczyk, T.; Śliwiński, T.; Skała, E. An In Vitro Estimation of the Cytotoxicity and Genotoxicity of Root Extract from Leonurus sibiricus L. Overexpressing AtPAP1 against Different Cancer Cell Lines. Molecules 2018, 23, 2049. https://doi.org/10.3390/molecules23082049
Sitarek P, Synowiec E, Kowalczyk T, Śliwiński T, Skała E. An In Vitro Estimation of the Cytotoxicity and Genotoxicity of Root Extract from Leonurus sibiricus L. Overexpressing AtPAP1 against Different Cancer Cell Lines. Molecules. 2018; 23(8):2049. https://doi.org/10.3390/molecules23082049
Chicago/Turabian StyleSitarek, Przemysław, Ewelina Synowiec, Tomasz Kowalczyk, Tomasz Śliwiński, and Ewa Skała. 2018. "An In Vitro Estimation of the Cytotoxicity and Genotoxicity of Root Extract from Leonurus sibiricus L. Overexpressing AtPAP1 against Different Cancer Cell Lines" Molecules 23, no. 8: 2049. https://doi.org/10.3390/molecules23082049





