Voltage-Dependent Anion Channel 2 of Arabidopsis thaliana (AtVDAC2) Is Involved in ABA-Mediated Early Seedling Development
Abstract
:1. Introduction
2. Results and Discussion
2.1. ABA Suppressed the Accumulation of AtVDAC2 Transcripts
2.2. Regulation of AtVDAC2 Promoter by ABA in the Protoplast Expression System
2.3. Generation of Sense and Antisense AtVDAC2 Transgenic Lines
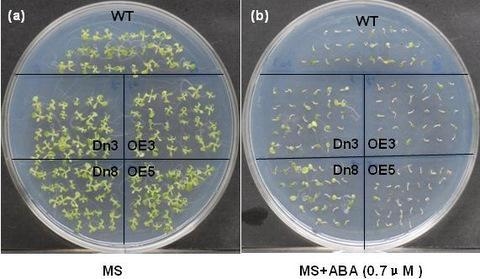
2.4. The AtVDAC2 Antisense Plants Shown an ABA-Insensitivity Phenotype during the Early Seedling Development in Arabidopsis
3. Experimental Section
3.1. Plant Material and Growth Conditions
3.2. Construction of Expression Vectors and Isolation of Transgenic Plants
3.3. Isolation of Arabidopsis Mesophyll Protoplasts
3.4. RNA Isolation and Semi-Quantitative RT-PCR
3.5. Transient Gene Expression in Arabidopsis Protoplasts
3.6. Phenotype Analysis of the AT VDAC2 Transgenic Plants
4. Conclusions
Acknowledgments
References and Notes
- Roosens, N; Al, BF; Jacobs, M; Homble, F. Characterization of a cDNA encoding a rice mitochondrial voltage-dependent anion channel and its gene expression studied upon plant development and osmotic stress. BBA-Biomembranes 2000, 1463, 470–476. [Google Scholar]
- Colombini, M. The Mitochondrial Voltage-Dependent Anion-Selective Channel. Biomembrane Electrochemistry 1994, 235, 245–258. [Google Scholar]
- Salinas, T; Duchene, AM; Delage, L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 18362–18367. [Google Scholar]
- Menard, G; Evrard, B; Bureau, M; Trottier, S. Cerebral Distribution of the B-36 Vdac Protein in Rat, Cow and Man Brain - Immunocytochemical Study. Cell Mol. Biol 1994, 40, 295–300. [Google Scholar]
- Lee, AC; Xu, X; Blachly-Dyson, E; Forte, M; Colombini, M. The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J. Membr. Biol 1998, 161, 173–181. [Google Scholar]
- Godbole, A; Varghese, J; Sarin, A; Mathew, MK. VDAC is a conserved element of death pathways in plant and animal systems. BBA-Mol. Cell Res 2003, 1642, 87–96. [Google Scholar]
- Galganska, H; Budzinska, M; Wojtkowska, M; Kmita, H. Redox regulation of protein expression in Saccharomyces cerevisiae mitochondria: Possible role of VDAC. Arch. Biochem. Biophys 2008, 479, 39–45. [Google Scholar]
- Lemasters, JJ; Holmuhamedov, E. Voltage-dependent anion channel (VDAC) as mitochondrial governator-Thinking outside the box. BBA-Mol. Basis Dis 2006, 1762, 181–190. [Google Scholar]
- Al Bitar, F; Roosens, N; Smeyers, M; Vauterin, M; Van, BJ; Jacobs, M; Homble, F. Sequence analysis, transcriptional and posttranscriptional regulation of the rice vdac family. BBA-Gene Struct. Expr 2003, 1625, 43–51. [Google Scholar]
- Elkeles, A; Breiman, A; Zizi, M. Functional differences among wheat voltage-dependent anion channel (VDAC) isoforms expressed in yeast - Indication for the presence of a novel VDAC-modulating protein? J. Biol. Chem 1997, 272, 6252–6260. [Google Scholar]
- Wang, J; Zhang, LD; Zuo, KJ; Oian, HM; Cao, YF; Tang, KX. Cloning and expressional studies of the voltage-dependent anion channel gene from Brassica rapa L. J. Integr. Plant Biol 2006, 48, 197–203. [Google Scholar]
- Tateda, C; Yamashita, K; Takahashi, F; Kusano, T; Takahashi, Y. Plant voltage-dependent anion channels are involved in host defense against Pseudomonas cichorii and in Bax-induced cell death. Plant Cell Rep 2009, 28, 41–51. [Google Scholar]
- Lacomme, C; Roby, D. Identification of new early markers of the hypersensitive response in Arabidopsis thaliana. FEBS Lett 1999, 459, 149–153. [Google Scholar]
- Desai, MK; Mishra, RN; Verma, D; Nair, S; Sopory, SK; Reddy, MK. Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum). Plant Physiol. Biochem 2006, 44, 483–493. [Google Scholar]
- Himmelbach, A; Yang, Y; Grill, E. Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol 2003, 6, 470–479. [Google Scholar]
- Christmann, A; Moes, D; Himmelbach, A; Yang, Y; Tang, Y; Grill, E. Integration of abscisic acid signalling into plant responses. Plant Biol 2006, 8, 314–325. [Google Scholar]
- Nambara, E; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol 2005, 56, 165–185. [Google Scholar]
- Finkelstein, RR; Gampala, SSL; Rock, CD. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar]
- Yoo, SD; Cho, YH; Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc 2007, 2, 1565–1572. [Google Scholar]
- Yang, Y. Signal transduction of abscisic acid in Arabidopsis thaliana: Identification and characterisation of protein interaction partners of ABI2. 2003.
- Li, YH; Lee, KK; Walsh, S; Smith, C; Hadingham, S; Sorefan, K; Cawley, G. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 2006, 16, 414–427. [Google Scholar]
- Lee, S; Leung, HT; Kim, E; Jang, J; Lee, E; Baek, K; Pak, WL. Effects of a mutation in the Drosophila porin gene encoding mitochondrial voltage-dependent anion channel protein on phototransduction. Dev. Neurobiol 2007, 67, 1533–1545. [Google Scholar]
- Abu-Hamad, S; Zaid, H; Israelson, A; Nahon, E; Shoshan-Barmatz, V. Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1 - Mapping the site of binding. J. Biol. Chem 2008, 283, 13482–13490. [Google Scholar]
- Huizing, M; Ruitenbeek, W; Thinnes, FP; DePinto, V; Wendel, U; Trijbels, FJM; Smit, LME. Deficiency of the voltage-dependent anion channel: A novel cause of mitochondriopathy. Pediatr. Res 1996, 39, 760–765. [Google Scholar]
- Abu-Hamad, S; Sivan, S; Shoshan-Barmatz, V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc. Natl. Acad. Sci. USA 2006, 103, 5787–5792. [Google Scholar]
- Dekkers, BJW; Schuurmans, JAMJ; Smeekens, SCM. Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis. Plant Mol. Biol 2008, 67, 151–167. [Google Scholar]
- Azoulay-Zohar, H; Israelson, A; Abu-Hamad, S; Shoshan-Barmatz, V. In self-defence: Hexokinase promotes voltage-dependent anion channel closure and prevents mitochondria-mediated apoptotic cell death. Biochem. J 2004, 377, 347–355. [Google Scholar]
- Zaid, H; Abu-Hamad, S; Israelson, A; Nathan, I; Shoshan-Barmatz, V. The voltage-dependent anion channel-1 modulates apoptotic cell death. Cell Death Differ 2005, 12, 751–760. [Google Scholar]
- Price, J; Li, TC; Kang, SG; Na, JK; Jang, JC. Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol 2003, 132, 1424–1438. [Google Scholar]
- Gincel, D; Zaid, H; Shoshan-Barmatz, V. Calcium binding and translocation by the voltage-dependent anion channel: A possible regulatory mechanism in mitochondrial function. Biochem. J 2001, 358, 147–155. [Google Scholar]
- Trewavas, AJ; Malho, R. Ca2+ signaling in plant cells: the big network! Cur. Opin. Plant Biol 1998, 1, 428–433. [Google Scholar]
- Zhang, XR; Henriques, R; Lin, SS; Niu, QW; Chua, NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc 2006, 1, 641–646. [Google Scholar]





© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yan, J.; He, H.; Tong, S.; Zhang, W.; Wang, J.; Li, X.; Yang, Y. Voltage-Dependent Anion Channel 2 of Arabidopsis thaliana (AtVDAC2) Is Involved in ABA-Mediated Early Seedling Development. Int. J. Mol. Sci. 2009, 10, 2476-2486. https://doi.org/10.3390/ijms10062476
Yan J, He H, Tong S, Zhang W, Wang J, Li X, Yang Y. Voltage-Dependent Anion Channel 2 of Arabidopsis thaliana (AtVDAC2) Is Involved in ABA-Mediated Early Seedling Development. International Journal of Molecular Sciences. 2009; 10(6):2476-2486. https://doi.org/10.3390/ijms10062476
Chicago/Turabian StyleYan, Jinping, Han He, Shibo Tong, Wanrong Zhang, Jianmei Wang, Xufeng Li, and Yi Yang. 2009. "Voltage-Dependent Anion Channel 2 of Arabidopsis thaliana (AtVDAC2) Is Involved in ABA-Mediated Early Seedling Development" International Journal of Molecular Sciences 10, no. 6: 2476-2486. https://doi.org/10.3390/ijms10062476