Co-localization of Carbonic Anhydrase and Phosphoenol-pyruvate Carboxylase and Localization of Pyruvate Kinase in Roots and Hypocotyls of Etiolated Glycine max Seedlings
Abstract
:1. Introduction
2. Results
2.1. CA activity and histochemical localization of CA activity in etiolated G. max seedlings
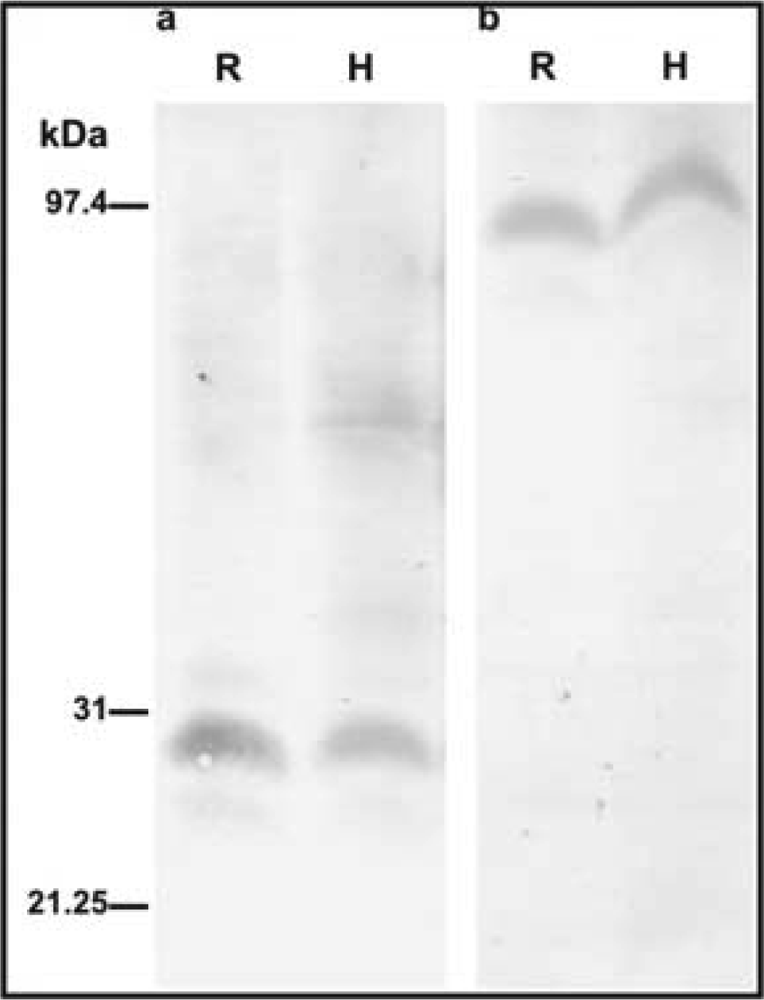
2.2. Immunohistochemical detection of CA and PEPC protein
2.3. Spatial patterns of GmCA1, GmPEPC7 and GmPK3 transcripts in primary and lateral roots of etiolated G. max seedlings
3. Discussion
4. Conclusions
5. Experimental Section
5.1. Plant material and growth conditions
5.2. Characterization of a cDNA clone coding for GmPK3
5.3. RT-PCR analysis of GmPK3 and GmPEPC7 transcript levels
5.4. In situ hybridization
5.5. Histochemical localization of CA activity
5.6. Total CA activity of G. max organ extracts
5.7. Protein extraction from G. max tissues and Western blot of PEPC and CA
5.8. Immunohistochemical detection of PEPC and CA
References and Notes
- Tripp, BC; Smith, K; Ferry, JG. Carbonic anhydrase: New insights for an ancient enzyme. J. Biol. Chem 2001, 276, 48615–48618. [Google Scholar]
- Lane, TW; Morel, FM. Regulation of carbonic anhydrase expression by zinc, cobalt, and carbon dioxide in the marine diatom Thalassiosira weissflogii. Plant Physiol 2000, 123, 345–352. [Google Scholar]
- So, AK-C; Aspie, GS; Williams, EB; Sively, JM; Heinhorst, S; Cannon, GC. A novel evolutionary lineage of carbonic anhydrase (ɛ class) is a component of the carboxysome shell. J. Bacteriol 2004, 186, 623–630. [Google Scholar]
- Sawaya, MR; Cannon, GC; Heinhorst, S; Tanaka, S; Williams, EB; Yeates, TO; Kerfeld, CA. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J. Biol. Chem 2006, 281, 7546–7555. [Google Scholar]
- Rowlett, RS; Tu, C; Murray, PS; Chamberlin, JE. Examination of the role of Gln-158 in the mechanism of CO2 hydration catalyzed by beta-carbonic anhydrase from Arabidopsis thaliana. Arch. Biochem. Biophys 2004, 425, 25–32. [Google Scholar]
- Parisi, G; Perales, M; Fornasari, MS; Colaneri, A; Gonzalez-Schain, N; Gomez-Casati, D; Zimmermann, S; Brennicke, A; Araya, A; Ferry, JG; Echave, J; Zabaleta, E. Gamma carbonic anhydrases in plant mitochondria. Plant Mol. Biol 2004, 55, 193–207. [Google Scholar]
- Fabre, N; Reiter, IM; Becuwe-Linka, N; Genty, B; Rumeau, D. Characterization and expression analysis of genes encoding alpha and beta carbonic anhydrases in Arabidopsis. Plant Cell Environ 2007, 30, 617–629. [Google Scholar]
- Moroney, JV; Bartlett, SG; Samuelsson, G. Carbonic anhydrases in plants and algae. Plant Cell Environ 2001, 24, 141–153. [Google Scholar]
- Price, GD; von Caemmerer, S; Evans, JR; Yu, JW; Lloyd, J; Oja, V; Kell, P; Harrison, K; Gallagher, A; Badger, MR. Specific reduction of chloroplast carbonic anhydrase activity by antisense RNA in transgenic tobacco has a minor effect on photosynthetic CO2 assimilation. Planta 1994, 193, 331–340. [Google Scholar]
- Jebanathirajah, JA; Coleman, JR. Association of carbonic anhydrase with a Calvin cycle enzyme complex in Nicotiana tabacum. Planta 1998, 204, 177–182. [Google Scholar]
- Burnell, JN; Hatch, MD. Low bundle sheath carbonic anhydrase is apparently essential for effective C4 pathway operation. Plant Physiol 1988, 86, 1252–1256. [Google Scholar]
- Chollet, R; Vidal, J; O’Leary, MH. Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol 1996, 47, 273–298. [Google Scholar]
- Von Caemmerer, S; Quinn, V; Hancock, NC; Price, GD; Furbank, RT; Ludwig, M. Carbonic anhydrase and C4 photosynthesis: A transgenic analysis. Plant Cell Environ 2004, 27, 697–703. [Google Scholar]
- Sánchez, R; Cejudo, FJ. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol 2003, 132, 949–957. [Google Scholar]
- Huppe, HC; Turpin, DH. Integration of carbon and nitrogen metabolism in plant and algal cells. Annu. Rev. Plant Physiol. Plant Mol. Biol 1994, 45, 577–607. [Google Scholar]
- Kavroulakis, N; Flemetakis, E; Aivalakis, G; Katinakis, P. Carbon metabolism in developing soybean root nodules: The role of carbonic anhydrase. Mol. Plant Microbe Interact 2000, 13, 14–22. [Google Scholar]
- Flemetakis, E; Dimou, M; Cotzur, D; Aivalakis, G; Efrose, RC; Kenoutis, C; Udvardi, M; Katinakis, P. A Lotus japonicus beta-type carbonic anhydrase gene expression pattern suggests distinct physiological roles during nodule development. Biochim Biophys. Acta 2003, 1628, 186–194. [Google Scholar]
- González, MC; Sanchez, R; Cejudo, FJ. Abiotic stresses affecting water balance induce phosphoenolpyruvate carboxylase expression in roots of wheat seedlings. Planta 2003, 216, 985–992. [Google Scholar]
- Sullivan, S; Jenkins, GI; Nimmo, HG. Roots, Cycles and Leaves. Expression of the Phosphoenolpyruvate Carboxylase Kinase Gene Family in Soybean. Plant Physiol 2004, 135, 2078–2087. [Google Scholar]
- Svensson, P; Blasing, OE; Westhoff, P. Evolution of C4 phosphoenolpyruvate carboxylase. Arch. Biochem. Biophys 2003, 414, 180–188. [Google Scholar]
- Raven, JA; Newman, JR. Requirement for carbonic anhydrase activity in processes other than photosynthetic inorganic carbon assimilation. Plant Cell Environ 1994, 17, 123–130. [Google Scholar]
- Coba de la Peña, T; Frugier, F; McKhann, HI; Bauer, P; Brown, S; Kondorosi, A; Crespi, MA. Carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobium during development. Plant J 1997, 11, 407–420. [Google Scholar]
- Aivalakis, G; Dimou, M; Flemetakis, E; Plati, F; Katinakis, P; Drossopoulos, JB. Immunolocalization of carbonic anhydrase and PEPC in developing seeds of Medicago sativa. Plant Physiol. Biochem 2004, 42, 181–186. [Google Scholar]
- Lance, C; Rustin, P. The central role of malate in plant metabolism. Physiol Veg 1984, 22, 625–641. [Google Scholar]
- Plaxton, WC. The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol 1996, 47, 185–214. [Google Scholar]
- Turner, WL; Plaxton, WC. Purification and characterization of cytosolic pyruvate kinase from banana fruit. Biochem J 2000, 352, 875–882. [Google Scholar]
- Chang, K; Roberts, JKM. Quantitation of rates of transport, metabolic fluxes, and cytoplasmic levels of inorganic carbon in maize root tips during K+ ion uptake. Plant Physiol 1992, 99, 291–297. [Google Scholar]
- Uhde-Stone, C; Gilbert, G; Johnson, JM-F; Litjens, R; Zinn, KE; Temple, SJ; Vance, CP; Allan, DL. Acclimation of white lupin to phosphorus deficiency involves enhanced expression of genes related to organic acid metabolism. Plant Soil 2003, 248, 99–116. [Google Scholar]
- Demir, N; Demir, Y; Yildirim, A. Carbonic anhydrases from leaves and roots of Daucus carota. Phytochemistry 1997, 44, 1247–1250. [Google Scholar]
- Tetu, SG; Tanz, SK; Vella, N; Burnell, JN; Ludwig, M. The Flaveria bidentis β-Carbonic anhydrase gene family encodes cytosolic and chloroplastic isoforms demonstrating distinct organ-specific expression patterns. Plant Physiol 2007, 144, 1316–1327. [Google Scholar]
- Majeau, N; Coleman, JR. Correlation of carbonic anhydrase and ribulose-1,5-bisphosphate carboxylase/oxygenase expression in pea. Plant Physiol 1994, 104, 1393–1399. [Google Scholar]
- Kavroulakis, N; Flemetakis, E; Aivalakis, G; Dahiya, P; Brewin, NJ; Fasseas, K; Hatzopoulos, P; Katinakis, P. Tissue distribution and subcellular localization of carbonic anhydrase in mature soybean root nodules indicates a role in CO2 diffusion. Plant Physiol. Biochem 2003, 41, 479–484. [Google Scholar]
- van der Westhuizen, MM; Cramer, MD. The influence of elevated root-zone dissolved inorganic carbon concentrations on respiratory O2 and CO2 flux in tomato roots. J. Exp. Bot 1998, 49, 1977–1985. [Google Scholar]
- Viktor, A; Cramer, MD. The influence of root assimilated inorganic carbon on nitrogen acquisition/assimilation and carbon partitioning. New Phytol 2005, 165, 157–169. [Google Scholar]
- Zeeman, SC; up Rees, T. Changes in carbohydrate metabolism and assimilate export starch-excess mutants of Arabidopsis. Plant Cell Environ 1999, 22, 1445–1453. [Google Scholar]
- Hibberd, JM; Quick, WP. Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 2002, 415, 451–454. [Google Scholar]
- Birnbaum, K; Shasha, DE; Wang, JY; Jung, JW; Lambert, GM; Galbraith, DW; Benfey, PN. A gene expression map of the Arabidopsis root. Science 2003, 302, 1956–1960. [Google Scholar]
- Drew, MC. Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol 1997, 48, 223–250. [Google Scholar]
- van Dongen, JT; Schurr, U; Pfister, M; Geigenberger, P. Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol 2003, 131, 1529–1543. [Google Scholar]
- Armstrong, W; Beckett, PM; Justin, SHFW; Lythe, S. Modeling and other aspects of root aeration by diffusion. In Plant Life under Oxygen Deprivation; Jackson, MB, Davies, DD, Eds.; SPB Academic Publishing: The Hague, The Netherlands, 1991; pp. 267–282. [Google Scholar]
- Gálvez, S; Hirsch, AM; Wycoff, KL; Hunt, S; Layzell, DB; Kondorosi, A; Crespi, M. Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol 2000, 124, 1059–1068. [Google Scholar]
- Dieuaide-Noubhani, M; Raffard, G; Canioni, P; Pradet, A; Raymond, P. Quantification of compartmented metabolic fluxes in maize root tips using isotope distribution from 13C- or 14C-labeled glucose. J. Biol. Chem 1995, 270, 13147–13159. [Google Scholar]
- Katz, J; Rognstad, R. The labeling of pentose phosphate from glucose-14C and estimation of the rates of transaldolase, transketolase, the contribution of the pentose cycle, and ribose phosphate synthesis. Biochemistry 1967, 6, 2227–2247. [Google Scholar]
- Adams, CA; Broman, TH; Rinne, RW. Use of [3,4-14C] glucose to assess in vivo competition for phosphoenolpyruvate between phosphoenolpyruvate carboxylase and pyruvate kinase in developing soybean seeds. Plant Cell Phyiol 1982, 23, 959–965. [Google Scholar]
- Rontein, D; Dieuaide-Noubhani, M; Dufourc, EJ; Raymond, P; Rolin, D. The metabolic architecture of plant cells. Stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J Biol Chem 2002, 277, 43948–43960. [Google Scholar]
- Dieuaide-Noubhani, M; Canioni, P; Raymond, P. Sugar-starvation-induced changes of carbon metabolism in excised maize root tips. Plant Physiol 1997, 115, 1505–1513. [Google Scholar]
- Plaxton, WC. The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol 1996, 47, 185–214. [Google Scholar]
- Altschul, SF; Madden, TL; Schaffer, AA; Zhang, J; Zhang, Z; Miller, W; Lipman, DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Brusslan, JA; Tobin, EM. Light-independent developmental regulation of cab gene expression in Arabidopsis thaliana seedlings. Proc. Natl. Acad. Sci. USA 1992, 89, 7791–7795. [Google Scholar]
- Dimou, M; Flemetakis, E; Delis, C; Aivalakis, G; Spyropulos, KG; Katinakis, P. Co-expression of two sugar transporters in primary and lateral roots of etiolated Glycine max seedlings. Plant Growth Regul 2005, 35, 259–266. [Google Scholar]
- Khalifah, RG. The carbon dioxide hydration activity of carbonic anhydrase. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem 1971, 246, 2561–2573. [Google Scholar]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]


| CA activitiy [μmole H+min−1 (mg protein)−1] | ||
|---|---|---|
| plumule | hypocotyl | root |
| ND (a) | 85.527±8.785 (b) | 216.363±51.969 (c) |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dimou, M.; Paunescu, A.; Aivalakis, G.; Flemetakis, E.; Katinakis, P. Co-localization of Carbonic Anhydrase and Phosphoenol-pyruvate Carboxylase and Localization of Pyruvate Kinase in Roots and Hypocotyls of Etiolated Glycine max Seedlings. Int. J. Mol. Sci. 2009, 10, 2896-2910. https://doi.org/10.3390/ijms10072896
Dimou M, Paunescu A, Aivalakis G, Flemetakis E, Katinakis P. Co-localization of Carbonic Anhydrase and Phosphoenol-pyruvate Carboxylase and Localization of Pyruvate Kinase in Roots and Hypocotyls of Etiolated Glycine max Seedlings. International Journal of Molecular Sciences. 2009; 10(7):2896-2910. https://doi.org/10.3390/ijms10072896
Chicago/Turabian StyleDimou, Maria, Anca Paunescu, Georgios Aivalakis, Emmanouil Flemetakis, and Panagiotis Katinakis. 2009. "Co-localization of Carbonic Anhydrase and Phosphoenol-pyruvate Carboxylase and Localization of Pyruvate Kinase in Roots and Hypocotyls of Etiolated Glycine max Seedlings" International Journal of Molecular Sciences 10, no. 7: 2896-2910. https://doi.org/10.3390/ijms10072896




