Photonic Methods to Enhance Fluorescence Correlation Spectroscopy and Single Molecule Fluorescence Detection
Abstract
:1. Introduction
2. Improved Single Molecule Fluorescence Detection by Structuring the Laser Excitation Beam
2.1. Total Internal Reflection Fluorescence Microscopy: TIRF
2.2. Fluorescence Detection on a Mirror
2.3. 4Pi Microscopy
2.4. Stimulated Emission Depletion Microscopy: STED
3. Improved Single Molecule Fluorescence Detection by Using Photonic Structures
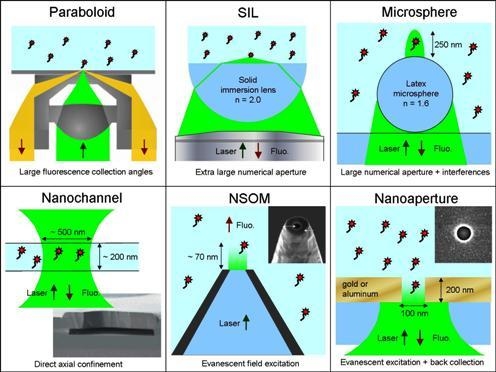
3.1. Paraboloid Collector
3.2. Solid Immersion Lens: SIL
3.3. Microspheres
3.4. Nanofluidic Channels
3.5. Near-Field Scanning Optical Microscope: NSOM
3.6. Nanometric Apertures
4. Conclusions
Acknowledgments
References
- Zander, C; Enderlein, J; Keller, RA. Single-Molecule Detection in Solution—Methods and Applications; VCH-Wiley: Berlin, Germany, 2002. [Google Scholar]
- Craighead, HG. Future lab-on-a-chip technologies for interrogating individual molecules. Nature 2006, 442, 387–393. [Google Scholar]
- Maiti, S; Haupts, U; Webb, WW. Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc. Nat. Acad. Sci. USA 1997, 94, 11753–11757. [Google Scholar]
- Webb, WW. Fluorescence correlation spectroscopy: Inception, biophysical experimentations, and prospectus. Appl. Opt 2001, 40, 3969–3983. [Google Scholar]
- Briddon, SJ; Hill, SJ. Pharmacology under the microscope: The use of fluorescence correlation spectroscopy to determine the properties of ligandreceptor complexes. Trends Pharmacol. Sci 2007, 28, 637–645. [Google Scholar]
- Huser, T. Nano-Biophotonics: New tools for chemical nano-analytics. Curr. Opin. Chem. Biol 2008, 12, 497–504. [Google Scholar]
- Blom, H; Kastrup, L; Eggeling, C. Fluorescence fluctuation spectroscopy in reduced detection Volumes. Curr. Pharm. Biotechnol 2006, 7, 51–66. [Google Scholar]
- Mannion, JT; Craighead, HG. Nanofluidic structures for single biomolecule fluorescent detection. Biopolymers 2006, 85, 131–143. [Google Scholar]
- Thompson, NL; Burghardt, TP; Axelrod, D. Measuring surface dynamics of biomolecules by total internal–Reflection fluorescence with photobleaching recovery or correlation spectroscopy. Biophys. J 1981, 33, 435–454. [Google Scholar]
- Hassler, K; Leutenegger, M; Rigler, P; Rao, R; Rigler, R; Gösch, M; Lasser, T. Total internal reflection fluorescence correlation spectroscopy (TIR-FCS) with low background and high count-rate per molecule. Opt. Express 2005, 13, 7415–7423. [Google Scholar]
- Starr, TE; Thompson, NL. Total internal reflection with fluorescence correlation spectroscopy: Combined surface reaction and solution diffusion. Biophys. J 2001, 80, 1575–1584. [Google Scholar]
- Borejdo, J; Calander, N; Gryczynski, Z; Gryczynski, I. Fluorescence correlation spectroscopy in surface plasmon coupled emission microscope. Opt. Express 2006, 14, 7878–7888. [Google Scholar]
- Leutenegger, M; Blom, H; Widengren, J; Eggeling, C; Gösch, M; Leitgeb, RA; Lasser, T. Dual-color total internal reflection fluorescence cross-correlation spectroscopy. J Biomed Optics 2006, 11, 040502. [Google Scholar]
- Lenne, PF; Etienne, E; Rigneault, H. Subwavelength patterns and high detection efficiency in fluorescence correlation spectroscopy using photonic structures. Appl. Phys. Lett 2002, 80, 4106–4108. [Google Scholar]
- Rigneault, H; Lenne, PF. Fluorescence correlation spectroscopy on a mirror. J. Opt. Soc. Am. B 2003, 20, 2203–2214. [Google Scholar]
- Etienne, E; Lenne, PF; Sturgis, JN; Rigneault, H. Confined diffusion in tubular structures analyzed by fluorescence correlation spectroscopy on a mirror. Appl. Opt 2006, 45, 4497–4507. [Google Scholar]
- Hell, SW. Toward fluorescence nanoscopy. Nat. Biotechnol 2003, 21, 1347–1355. [Google Scholar]
- Hell, SW; Stelzer, EHK. Fundamental improvement of resolution with a 4Pi-confocal fluorescence microscope using twophoton excitation. Opt. Commun 1992, 93, 277–282. [Google Scholar]
- Arkhipov, A; Hüve, J; Kahms, M; Peters, R; Schulten, K. Continuous fluorescence microphotolysis and correlation spectroscopy using 4Pi microscopy. Biophys. J 2007, 93, 4006–4017. [Google Scholar]
- Kastrup, L; Blom, H; Eggeling, C; Hell, SW. Fluorescence fluctuation spectroscopy in subdiffraction focal volumes. Phys Rev Lett 2005, 94, 178104:1–178104:4. [Google Scholar]
- Eggeling, C; Ringemann, C; Medda, R; Schwarzmann, G; Sandhoff, K; Polyakova, S; Belov, VN; Hein, B; von Middendorff, C; Schönle, A; Hell, SW. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 2008, 457, 1159–1162. [Google Scholar]
- Enderlein, J; Ruckstuhl, T; Seeger, S. Highly efficient optical detection of surface-generated fluorescence. Appl. Opt 1999, 38, 724–732. [Google Scholar]
- Ruckstuhl, T; Enderlein, J; Jung, S; Seeger, S. Forbidden light detection from single molecules. Anal. Chem 2000, 72, 2117–2123. [Google Scholar]
- Ries, J; Ruckstuhl, T; Verdes, D; Schwille, P. Supercritical angle fluorescence correlation spectroscopy. Biophys. J 2008, 94, 221–229. [Google Scholar]
- Koyama, K; Yoshita, M; Baba, M; Suemoto, T; Akiyama, H. High collection efficiency in fluorescence microscopy with a Solid Immersion Lens. Appl. Phys. Lett 1999, 75, 1667–1669. [Google Scholar]
- Serov, A; Rao, R; Gösch, M; Anhut, T; Martin, D; Brunner, R; Rigler, R; Lasser, T. High light field confinement for fluorescent correlation spectroscopy using a solid immersion lens. Biosens. Bioelectron 2004, 20, 431–435. [Google Scholar]
- Rao, R; Mitic, J; Serov, A; Leitgeb, RA; Lasser, T. Field confinement with aberration correction for solid immersion lens based fluorescence correlation spectroscopy. Opt. Commun 2007, 271, 462–469. [Google Scholar]
- Gérard, D; Wenger, J; Devilez, A; Gachet, D; Stout, B; Bonod, N; Popov, E; Rigneault, H. Strong electromagnetic confinement near dielectric microspheres to enhance single-molecule fluorescence. Opt. Express 2008, 16, 15297–15303. [Google Scholar]
- Devilez, A; Bonod, N; Stout, B; Gérard, D; Wenger, J; Rigneault, H; Popov, E. Three-dimensional subwavelength confinement of photonic nanojets. Opt. Express 2009, 17, 2089–2094. [Google Scholar]
- Gérard, D; Devilez, A; Aouani, H; Stout, B; Bonod, N; Wenger, J; Popov, E; Rigneault, H. Efficient excitation and collection of single molecule fluorescence close to a dielectric microsphere. J. Opt. Soc. Am. B 2009, 26, 1473–1478. [Google Scholar]
- Wenger, J; Gérard, D; Aouani, H; Rigneault, H. Disposable microscope objective lenses for fluorescence correlation spectroscopy using latex microspheres. Anal. Chem 2008, 80, 6800–6804. [Google Scholar]
- Aouani, H; Deiss, F; Wenger, J; Ferrand, P; Sojic, N; Rigneault, H. Optical-fiber-microsphere for remote fluorescence correlation spectroscopy. Opt. Express 2009, 17, 18912–18919. [Google Scholar]
- Brinkmeier, M; Dijrre, K; Riebeseel, K; Rigler, R. Confocal spectroscopy in microstructures. Biophys. Chem 1997, 66, 229–239. [Google Scholar]
- Foquet, M; Korlach, J; Zipfel, WR; Webb, WW; Craighead, HG. DNA fragment sizing by single molecule detection in submicrometer-sized closed fluidic channels. Anal. Chem 2002, 74, 1415–1422. [Google Scholar]
- Foquet, M; Korlach, J; Zipfel, WR; Webb, WW; Craighead, HG. Focal volume confinement by submicrometer-sized fluidic channels. Anal. Chem 2004, 76, 1618–1626. [Google Scholar]
- Lenne, PF; Colombo, D; Giovannini, H; Rigneault, H. Flow profiles and directionality in microcapillaries measured by fluorescence correlation spectroscopy. Single Mol 2002, 3, 194–200. [Google Scholar]
- Yin, D; Lunt, EJ; Barman, A; Hawkins, AR; Schmidt, H. Microphotonic control of single molecule fluorescence correlation spectroscopy using planar optofluidics. Opt. Express 2007, 15, 7290–7295. [Google Scholar]
- Lewis, A; Taha, H; Strinkovski, A; Menevitch, A; Katchatouriants, A; Dekhter, R; Amman, E. Near-field optics: From subwavelength illumination to nanometric shadowing. Nat. Biotechnol 2003, 21, 1378–1386. [Google Scholar]
- Vobornik, D; Banks, DS; Lu, Z; Fradin, C; Taylor, R; Johnston, LJ. Fluorescence correlation spectroscopy with sub-diffraction-limited resolution using near-field optical probes. Appl Phys Lett 2008, 93, 163904:1–163904:3. [Google Scholar]
- Lu, G; Lei, FH; Angiboust, JF; Manfait, M. Confined detection volume of fluorescence correlation spectroscopy by bare fiber probes. Eur Biophys J 2009. [Google Scholar]
- Herrmann, M; Neuberth, N; Wissler, J; Pérez, J; Gradl, D; Naber, A. Near-field optical study of protein transport kinetics at a single nuclear pore. Nano Lett 2009, 9, 3330–3336. [Google Scholar]
- Levene, MJ; Korlach, J; Turner, SW; Foquet, M; Craighead, HG; Webb, WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 2003, 299, 682–686. [Google Scholar]
- Samiee, KT; Foquet, M; Guo, L; Cox, EC; Craighead, HG. Lambda repressor oligomerization kinetics at high concentrations using fluorescence correlation spectroscopy in zero-mode waveguides. Biophys. J 2005, 88, 2145–2153. [Google Scholar]
- Leutenegger, M; Gösch, M; Perentes, A; Hoffmann, P; Martin, OJF; Lasser, T. Confining the sampling volume for Fluorescence Correlation Spectroscopy using a sub-wavelength sized aperture. Opt. Express 2006, 14, 956–969. [Google Scholar]
- Wenger, J; Gérard, D; Lenne, PF; Rigneault, H; Dintinger, J; Ebbesen, TW; Boned, A; Conchonaud, F; Marguet, D. Dual-color fluorescence cross-correlation spectroscopy in a single nanoaperture: Towards rapid multicomponent screening at high concentrations. Opt. Express 2006, 14, 12206–12216. [Google Scholar]
- Liao, D; Galajda, P; Riehn, R; Ilic, R; Puchalla, JL; Yu, HG; Craighead, HG; Austin, RH. Single molecule correlation spectroscopy in continuous flow mixers with zero-mode waveguides. Opt. Express 2008, 16, 10077–10090. [Google Scholar]
- Rigneault, H; Capoulade, J; Dintinger, J; Wenger, J; Bonod, N; Popov, E; Ebbesen, TW; Lenne, PF. Enhancement of single-molecule fluorescence detection in subwavelength apertures. Phys Rev Lett 2005, 95, 117401:1–117401:4. [Google Scholar]
- Wenger, J; Lenne, PF; Popov, E; Rigneault, H; Dintinger, J; Ebbesen, TW. Single molecule fluorescence in rectangular nano-apertures. Opt. Express 2005, 13, 7035–7044. [Google Scholar]
- Wenger, J; Gérard, D; Bonod, N; Popov, E; Rigneault, H; Dintinger, J; Mahboub, O; Ebbesen, TW. Emission and excitation contributions to enhanced single molecule fluorescence by gold nanometric apertures. Opt. Express 2008, 16, 3008–3020. [Google Scholar]
- Gérard, D; Wenger, J; Bonod, N; Popov, E; Rigneault, H; Mahdavi, F; Blair, S; Dintinger, J; Ebbesen, TW. Nanoaperture-enhanced fluorescence: Towards higher detection rates with plasmonic metals. Phys Rev B 2008, 77, 045413. [Google Scholar]
- Aouani, H; Wenger, J; Gérard, D; Rigneault, H; Devaux, E; Ebbesen, TW; Mahdavi, F; Xu, T; Blair, S. Crucial role of the adhesion layer on the plasmonic fluorescence enhancement. ACS Nano 2009, 3, 2043–2048. [Google Scholar]
- Wenger, J; Gérard, D; Aouani, H; Rigneault, H; Lowder, B; Blair, S; Devaux, E; Ebbesen, TW. Nanoaperture-enhanced signal-to-noise ratio in fluorescence correlation spectroscopy. Anal. Chem 2009, 81, 834–839. [Google Scholar]
- Lenne, PF; Rigneault, H; Marguet, D; Wenger, J. Fluorescence fluctuations analysis in nanoapertures: Physical concepts and biological applications. Histochem. Cell Biol 2008, 130, 795–805. [Google Scholar]
- Samiee, KT; Moran-Mirabal, JM; Cheung, YK; Craighead, HG. Zero mode waveguides for single-molecule spectroscopy on lipid membranes. Biophys. J 2006, 90, 3288–3299. [Google Scholar]
- Wenger, J; Rigneault, H; Dintinger, J; Marguet, D; Lenne, PF. Single-fluorophore diffusion in a lipid membrane over a subwavelength aperture. J Biol Phys 2006, 32, SN1–SN4. [Google Scholar]
- Edel, JB; Wu, M; Baird, B; Craighead, HG. High spatial resolution observation of single molecule dynamics in living cell membranes. Biophys. J 2005, 88, L43–L45. [Google Scholar]
- Wenger, J; Conchonaud, F; Dintinger, J; Wawrezinieck, L; Ebbesen, TW; Rigneault, H; Marguet, D; Lenne, PF. Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys. J 2007, 92, 913–919. [Google Scholar]
- Moran-Mirabal, JM; Torres, AJ; Samiee, KT; Baird, B; Craighead, HG. Cell investigation of nanostructures: Zero-mode waveguides for plasma membrane studies with single molecule resolution. Nanotechnology 2007, 18, 195101:1–195101:10. [Google Scholar]
- Eid, J; Fehr, A; Gray, J; Luong, K; Lyle, J; Otto, G; Peluso, P; Rank, D; Baybayan, P; Bettman, B; Bibillo, A; Bjornson, K; Chaudhuri, B; Christians, F; Cicero, R; Clark, S; Dalal, R; deWinter, A; Dixon, J; Foquet, M; Gaertner, A; Hardenbol, P; Heiner, C; Hester, K; Holden, D; Kearns, G; Kong, X; Kuse, R; Lacroix, Y; Lin, S; Lundquist, P; Ma, C; Marks, P; Maxham, M; Murphy, D; Park, I; Pham, T; Phillips, M; Roy, J; Sebra, R; Shen, G; Sorenson, J; Tomaney, A; Travers, K; Trulson, M; Vieceli, J; Wegener, J; Wu, D; Yang, A; Zaccarin, D; Zhao, P; Zhong, F; Korlach, J; Turner, S. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar]





| Type | Volume reduction | Fluo. enhancement | Complexity |
|---|---|---|---|
| Confocal | x1 | x1 | * |
| TIRF | x10 | x2 | * |
| Mirror | x6 | x4 | * |
| 4Pi | x6 | x4 | *** |
| STED | x100 | ? | *** |
| Paraboloid | x10 | x2 | * |
| SIL | x3 | x1.5 | ** |
| Microsphere | x10 | x5 | * |
| Nanofluidics | x100 | x3 | ** |
| NSOM | x100 | ? | *** |
| Nanoapertures | x1000 | x25 | ** |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wenger, J.; Rigneault, H. Photonic Methods to Enhance Fluorescence Correlation Spectroscopy and Single Molecule Fluorescence Detection. Int. J. Mol. Sci. 2010, 11, 206-221. https://doi.org/10.3390/ijms11010206
Wenger J, Rigneault H. Photonic Methods to Enhance Fluorescence Correlation Spectroscopy and Single Molecule Fluorescence Detection. International Journal of Molecular Sciences. 2010; 11(1):206-221. https://doi.org/10.3390/ijms11010206
Chicago/Turabian StyleWenger, Jérome, and Hervé Rigneault. 2010. "Photonic Methods to Enhance Fluorescence Correlation Spectroscopy and Single Molecule Fluorescence Detection" International Journal of Molecular Sciences 11, no. 1: 206-221. https://doi.org/10.3390/ijms11010206