Microbial Contamination of Orthodontic Buccal Tubes from Manufacturers
Abstract
:1. Introduction
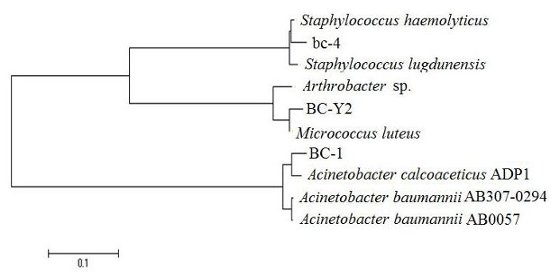
2. Results and Discussion
3. Materials and Methods
3.1. Buccal Tube Samples
3.2. Isolation of Microorganisms
3.3. DNA Extraction, PCR and Sequencing
3.4. Analysis of Bacterial 16S rRNA Sequences
4. Conclusions
Acknowledgement
References
- Kuramitsu, HK; He, X; Lux, R; Anderson, MH; Shi, W. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev 2007, 71, 653–670. [Google Scholar]
- Mazumdar, V; Snitkin, ES; Amar, S; Segre, D. Metabolic network model of a human oral pathogen. J. Bacteriol 2009, 191, 74–90. [Google Scholar]
- Spaulding, EH. Chemical disinfection and antisepsis in the hospitals. J. Hosp. Res 1972, 9, 5–31. [Google Scholar]
- Lucas, VS; Omar, J; Vieira, A; Roberts, GJ. The relationship between odontogenic bacteraemia and orthodontic treatment procedures. Eur. J. Orthod 2002, 24, 293–301. [Google Scholar]
- Miller, CH; Palenik, CJ. Infection Control and Management of Hazardous Materials for the Dental Team, 2nd ed; Mosby Building Arts: St. Louise, MO, USA, 1998. [Google Scholar]
- Wichelhaus, A; Bader, F; Sander, FG; Krieger, D; Mertens, T. Effective disinfection of orthodontic pliers. J. Orofac. Orthop 2006, 67, 316–336. [Google Scholar]
- Benson, PE; Douglas, CWI. Decontamination of orthodontic bands following size determination and cleaning. J. Orthod 2007, 34, 18–24. [Google Scholar]
- Brusca, MI; Chara, O; Sterin-Borda, L; Rosa, AC. Influence of different orthodontic brackets on adherence of microorganisms in vitro. Angle Orthod 2007, 77, 331–336. [Google Scholar]
- Papaioannou, W; Gizani, S; Nassika, M; Kontou, E; Nakou, M. Adhesion of Streptococcus mutans to different types of brackets. Angle Orthod 2007, 77, 1090–1095. [Google Scholar]
- Saitou, N; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol 1980, 16, 111–120. [Google Scholar]
- Tamura, K; Dudley, J; Nei, M; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol 2007, 24, 1596–1599. [Google Scholar]
- Sukontapatipark, W; El-Agroudi, MA; Selliseth, NJ; Thunold, K; Selvig, KA. Bacterial colonization associated with fixed orthodontic appliances: a scanning electron microscopy study. Eur. J. Orthod 2001, 23, 475–484. [Google Scholar]
- Pandis, N; Papaioannou, W; Kontou, E; Nakou, M; Makou, M; Eliades, T. Salivary Streptococcus mutans levels in patients with conventional and self-ligating brackets. Eur. J. Orthod 2010, 32, 94–99. [Google Scholar]
- Chiller, K; Selkin, BA; Murakawa, GJ. Skin microflora and bacterial infections of the skin. J. Investig. Dermatol. Symp. Proc 2001, 6, 170–174. [Google Scholar]
- Greenblatt, CL; Baum, J; Klein, BY; Nachshon, S; Koltunov, V; Cano, RJ. Micrococcus luteus survival in amber. Microbiol. Ecol 2004, 48, 120–127. [Google Scholar]
- Mukamolova, GV; Murzin, AG; Salina, EG; Demina, GR; Kell, DB; Kaprelyants, AS; Young, M. Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol. Microbiol 2006, 59, 84–98. [Google Scholar]
- Kocur, M; Kloos, WE; Schleifer, KH. The genus Micrococcus. In The Prokaryotes; Sringer-Verlag: New York, NY, USA, 1992; Volume II. [Google Scholar]
- Wharton, M; Rice, JR; McCallum, R; Gallis, HA. Septic arthritis due to Micrococcus luteus. J. Rheumatol 1986, 13, 659–660. [Google Scholar]
- Albertson, D; Natsios, GA; Gleckman, R. Septic shock with Micrococcus luteus. Arch. Intern. Med 1978, 138, 487–488. [Google Scholar]
- Fosse, T; Peloux, Y; Granthil, C; Toga, B; Bertrando, J; Sethian, M. Meningitis due to Micrococcus luteus. Infection 1985, 13, 280–281. [Google Scholar]
- Souhami, L; Feld, R; Tuffnell, PG; Feller, T. Micrococcus luteus pneumonia: a case report and review of the literature. Med. Pediatr. Oncol 1979, 7, 309–314. [Google Scholar]
- Peleg, AY; Seifert, H; Paterson, DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev 2008, 21, 538–582. [Google Scholar]
- Retailliau, HF; Hightower, AW; Dixon, RE; Allen, JR. A nosocomial pathogen with an unusual seasonal pattern. J. Infect. Dis 1979, 139, 371–375. [Google Scholar]
- Williams, HN; Falkler, WA, Jr; Hasler, JF. Acinetobacter contamination of laboratory dental pumice. J. Dent. Res 1983, 62, 1073–1075. [Google Scholar]
- Lina, G; Etienne, J; Vandenesch, F. Fischetti, VA, Novick, RP, Ferretti, JJ, Portnoy, DA, Rood, JI, Eds.; Biology and pathogenicity of staphylococci other then Staphylococcus aureus and Staphylococcus epidermidis. In Gram-Positive Pathogens; ASM Press: Washington, DC, USA, 2000; pp. 450–462. [Google Scholar]
- Pfaller, MA; Jones, RN; Doern, GV; Sader, HS; Kugler, KC; Beach, ML. Survey of blood stream infections attributable to gram-positive cocci: Frequency of occurrence and antimicrobial susceptibility of isolates collected in 1997 in the United States, Canada, and Latin America from the SENTRY Antimicrobial Surveillance Program. SENTRY Participants Group. Diagn. Microbiol. Infect. Dis 1999, 33, 283–297. [Google Scholar]
- Kjelleberg, S; Molin, S. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol 2002, 5, 254–258. [Google Scholar]
- Shittu, A; Lin, J; Morrison, D; Kolawole, D. Isolation and molecular characterization of multiresistant Staphylococcus sciuri and Staphylococcus haemolyticus associated with skin and soft-tissue infections. J. Med. Microbiol 2004, 53, 51–55. [Google Scholar]
- Getchell-White, SI; Donowitz, LG; Groschel, DH. The inanimate environment of an intensive care unit as a potential source of nosocomial bacteria: Evidence for long survival of Acinetobacter calcoaceticus. Infect. Control Hosp. Epidemiol 1989, 10, 402–407. [Google Scholar]
- Chan, KG; Tiew, SZ; Ng, CC. Rapid isolation method of soil bacilli and screening of their quorum quenching activity. As. Pac. J. Mol. Biol. Biotech 2007, 15, 153–156. [Google Scholar]
- Chan, KG; Yin, WF; Sam, CK; Koh, CL. A novel medium for the isolation of N-acylhomoserine lactone-degrading bacteria. J. Ind. Microbiol. Biotechnol 2009, 36, 247–251. [Google Scholar]

| Group | Type of Buccal | Tube Manufacturer | Prescription |
|---|---|---|---|
| A | Lower right molar single tube | American Othodontics, Sheboygan, Wis | MBT |
| B | Lower right molar single tube—small base | 3M Unitek, Monrovia Calif | MBT |
| C | Lower right molar single tube—large base | 3M Unitek, Monrovia Calif | MBT |
| D | Lower right molar single tube | Hangzhou Dentop, China | MBT |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Purmal, K.; Chin, S.; Pinto, J.; Yin, W.-F.; Chan, K.-G. Microbial Contamination of Orthodontic Buccal Tubes from Manufacturers. Int. J. Mol. Sci. 2010, 11, 3349-3356. https://doi.org/10.3390/ijms11093349
Purmal K, Chin S, Pinto J, Yin W-F, Chan K-G. Microbial Contamination of Orthodontic Buccal Tubes from Manufacturers. International Journal of Molecular Sciences. 2010; 11(9):3349-3356. https://doi.org/10.3390/ijms11093349
Chicago/Turabian StylePurmal, Kathiravan, Shenyang Chin, John Pinto, Wai-Fong Yin, and Kok-Gan Chan. 2010. "Microbial Contamination of Orthodontic Buccal Tubes from Manufacturers" International Journal of Molecular Sciences 11, no. 9: 3349-3356. https://doi.org/10.3390/ijms11093349