The Chitinolytic Activities of Streptomyces sp. TH-11
Abstract
:1. Introduction
2. Results and Discussion
2.1. Degradation of Chitin Powder by Streptomyces sp. TH-11
2.2. In-gel Activity Staining of Chitinolytic Enzymes from Streptomyces sp. TH-11
2.3. Decomposition of Shrimp Shells by Streptomyces sp. TH-11
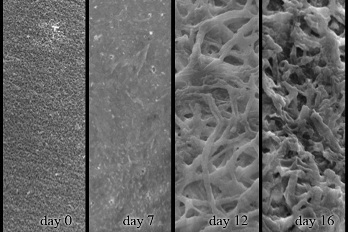
2.4. Decomposition of Shrimp Shells Visualized by SEM and AFM
3. Experimental Section
4. Conclusions
Acknowledgements
References
- Kumar, MN; Muzzarelli, RA; Muzzarelli, C; Sashiwa, H; Domb, AJ. Chitosan chemistry and pharmaceutical perspectives. Chem. Rev 2004, 104, 6017–6084. [Google Scholar]
- Liau, CY; Lin, CS. Detection of chitinolytic enzymes in Ipomoea batatas leaf extract by activity staining after gel electrophoresis. J. Chin. Chem. Soc 2008, 55, 678–681. [Google Scholar]
- Liau, CY; Lin, CS. A modified Coomassie Brilliant Blue G 250 staining method for the detection of chitinase activity and molecular weight after polyacrylamide gel electrophoresis. J. Biosci. Bioeng 2008, 106, 111–113. [Google Scholar]
- Chater, KF; Biro, S; Lee, KJ; Palmer, T; Schrempf, H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev 2009, 34, 171–198. [Google Scholar]
- Bentley, SD; Chater, KF; Cerdeño-Tárraga, AM; Challis, GL; Thomson, NR; James, KD; Harris, DE; Quail, MA; Kieser, H; Harper, D; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar]
- Kawase, T; Yokokawa, S; Saito, A; Fujii, T; Nikaidou, N; Miyashita, K; Watanabe, T. Comparison of Enzymatic and Antifungal Properties between Family 18 and 19 Chitinases from S. coelicolor A3(2). Biosci. Biotechnol. Biochem 2006, 70, 988–998. [Google Scholar]
- Hoang, KC; Lee, CY; Tseng, M; Chu, WC. Polyester-degrading actinomycetes isolated from the Touchien River of Taiwan. World J. Microbiol. Biotechnol 2007, 23, 201–205. [Google Scholar]
- Hoang, KC; Lee, CY; Lai, YC; Liau, CY. TH-11, a Streptomyces sp. strain that degrades poly(3-hydroxybutyrate) and poly(ethylene succinate). J. Chin. Chem. Soc 2008, 55, 1214–1220. [Google Scholar]
- Nawani, NN; Kapadnis, BP. Optimization of chitinase production using statistics based experimental designs. Process Biochem 2005, 40, 651–660. [Google Scholar]
- Hjort, K; Bergstrom, M; Adesina, MF; Jansson, JK; Smalla, K; Sjöling, S. Chitinase genes revealed and compared in bacterial isolates, DNA extracts and a metagenomic library from a phytopathogen-suppressive soil. FEMS Microbiol. Ecol 2010, 71, 197–207. [Google Scholar]
- Bhattacharya, D; Nagpure, A; Gupta, RK. Bacterial chitinases: properties and potential. Critical Rev. Biotechnol 2007, 27, 21–28. [Google Scholar]
- Narayana, KJP; Vijayalakshmi, M. Chitinase Production by Streptomyces sp. ANU 6277. Braz. J. Microbiol 2009, 40, 725–733. [Google Scholar]
- Kawase, T; Saito, A; Sato, T; Kanai, R; Fujii, T; Nikaidou, N; Miyashita, K; Watanabe, T. Distribution and Phylogenetic Analysis of Family 19 Chitinases in Actinobacteria. Appl. Environ. Micorbiol 2004, 70, 1135–1144. [Google Scholar]
- Percot, A; Viton, C; Domard, A. Optimization of chitin extraction from shrimp shells. Biomacromolecules 2003, 4, 12–18. [Google Scholar]
- Imoto, T; Yagishita, K. A simple activity measurement of lysozyme. Agric. Biol. Chem 1971, 35, 1154–1156. [Google Scholar]






© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hoang, K.-C.; Lai, T.-H.; Lin, C.-S.; Chen, Y.-T.; Liau, C.-Y. The Chitinolytic Activities of Streptomyces sp. TH-11. Int. J. Mol. Sci. 2011, 12, 56-65. https://doi.org/10.3390/ijms12010056
Hoang K-C, Lai T-H, Lin C-S, Chen Y-T, Liau C-Y. The Chitinolytic Activities of Streptomyces sp. TH-11. International Journal of Molecular Sciences. 2011; 12(1):56-65. https://doi.org/10.3390/ijms12010056
Chicago/Turabian StyleHoang, Kim-Chi, Tzu-Hsuan Lai, Chung-Sheng Lin, Ying-Tsong Chen, and Chun-Yi Liau. 2011. "The Chitinolytic Activities of Streptomyces sp. TH-11" International Journal of Molecular Sciences 12, no. 1: 56-65. https://doi.org/10.3390/ijms12010056