A Concise Review on Epigenetic Regulation: Insight into Molecular Mechanisms
Abstract
:1. Introduction
2. Transition to Pluripotency
3. Molecular Signals in Epigenetic Regulation
3.1. Main Epigenetic Regulatory Mechanisms
3.2. Transcriptional Regulation
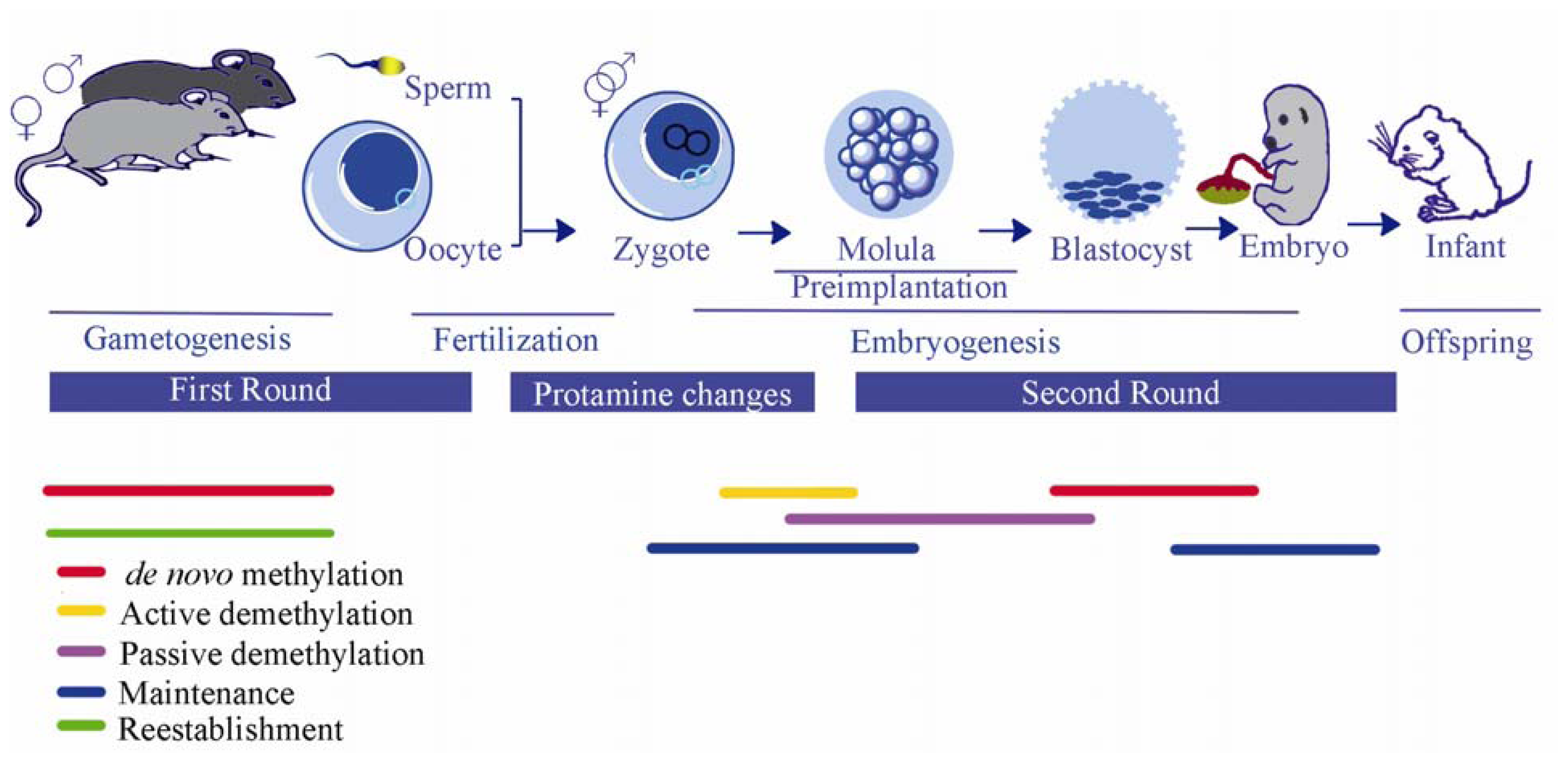
3.3. Epigenetic Reprogramming During Embryogenesis
3.4. Epigenetic Features of DNA Methylation
3.5. DNA Methylation Signals
3.6. DNA Methylation Analysis
3.7. Regulatory Factors in DNA Methylation
3.8. DNA Methyltransferases
3.9. Epigenetic Features of ncRNAs
3.10. Epigenetic Features of Small RNAs
3.11. Epigenetic Features of Chromatin Modifications
4. General Features of Imprinted Genes and Their Regulation
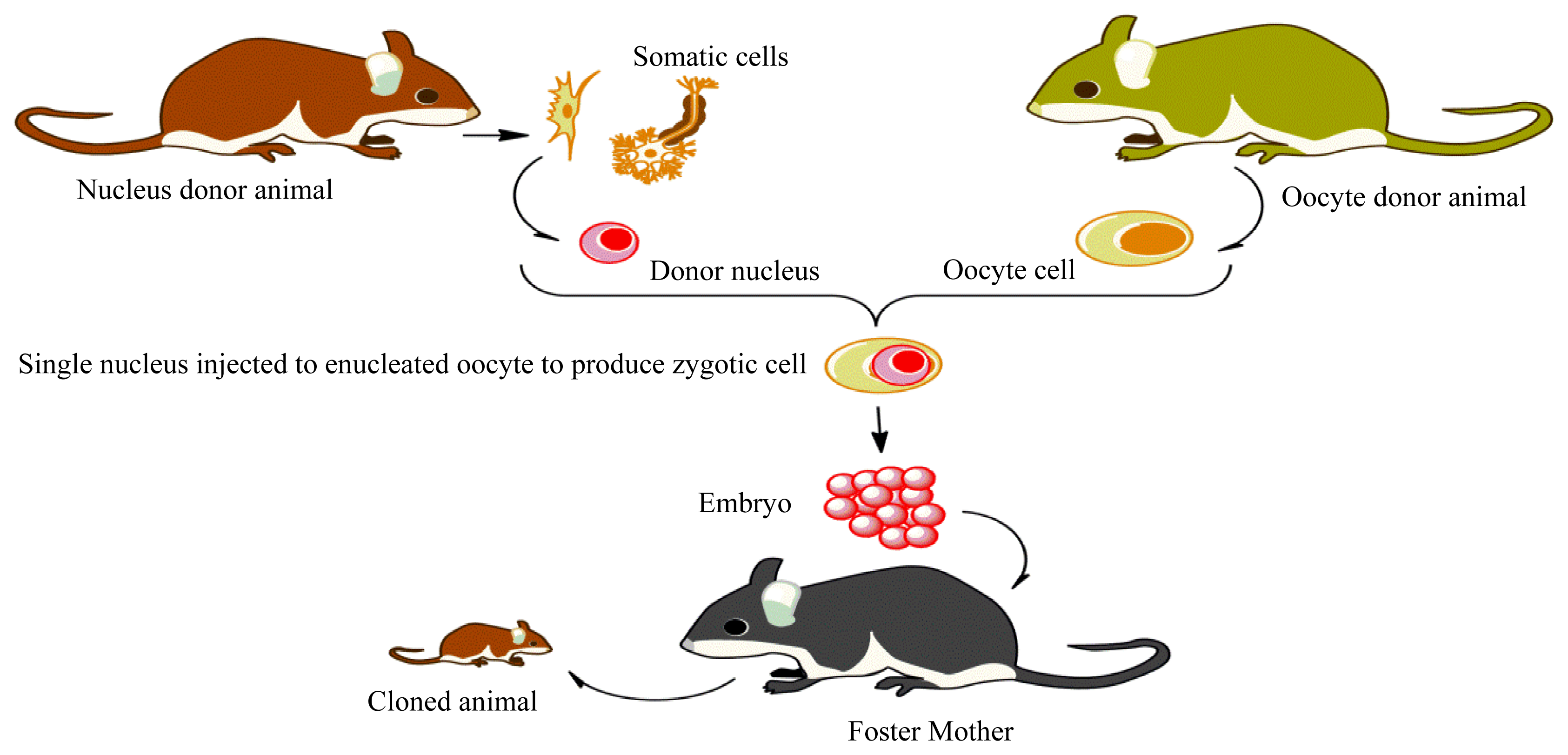
Methylation of Imprinted Genes and Its Abnormalities in Cloned Animals
5. Control of Gene Expression During Gametogenesis
6. Epigenetic Regulation During Gestation
7. Epigenetic Regulation During Embryogenesis
8. Epigenetic Regulation and Placental Development
9. Transcriptional Regulation by Polycomb Protein
10. Conclusion
Acknowledgments
References
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar]
- Mikkelsen, T.S.; Hanna, J.; Zhang, X.; Ku, M.; Wernig, M.; Schorderet, P.; Bernstein, B.E.; Jaenisch, R.; Lander, E.S.; Meissner, A. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008, 454, 49–55. [Google Scholar]
- Yang, X.; Smith, S.L.; Tian, X.C.; Lewin, H.A.; Renard, J.-P.; Wakayama, T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet 2007, 39, 295–302. [Google Scholar]
- Rideout, W.M.; Eggan, K.; Jaenisch, R. Nuclear cloning and epigenetic reprogramming of the genome. Science 2001, 293, 1093–1098. [Google Scholar]
- Keefer, C.L. Lessons learned from nuclear transfer (cloning). Theriogenology 2008, 69, 48–54. [Google Scholar]
- Yamanaka, S.; Blau, H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature 2010, 465, 704–712. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar]
- Pells, S.; Di Domenico, A.I.; Gallagher, E.J.; McWhir, J. Multipotentiality of neuronal cells after spontaneous fusion with embryonic stem cells and nuclear reprogramming in vitro. Cloning Stem Cells 2002, 4, 331–338. [Google Scholar]
- Tada, M.; Takahama, Y.; Abe, K.; Nakatsuji, N.; Tada, T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol 2001, 11, 1553–1558. [Google Scholar]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar]
- Yu, J.Y.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar]
- Bhutani, N.; Brady, J.J.; Damian, M.; Sacco, A.; Corbel, S.Y.; Blau, H.M. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 2010, 463, 1042–1047. [Google Scholar]
- Morgan, H.D.; Dean, W.; Coker, H.A.; Reik, W.; Petersen-Mahrt, S.K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues—Implications for epigenetic reprogramming. J. Biol. Chem 2004, 279, 52353–52360. [Google Scholar]
- Constant, F.; Guillomot, M.; Heyman, Y.; Vignon, X.; Laigre, P.; Servely, J.L.; Renard, J.P.; Chavatte-Palmer, P. Large offspring or large placenta syndrome? Morphometric analysis of late gestation bovine placentomes from somatic nuclear transfer pregnancies complicated by hydrallantois. Biol. Reprod 2006, 75, 122–130. [Google Scholar]
- Tamashiro, K.L.K.; Wakayama, T.; Blanchard, R.J.; Blanchard, D.C.; Yanagimachi, R. Postnatal growth and behavioral development of mice cloned from adult cumulus cells. Biol. Reprod 2000, 63, 328–334. [Google Scholar]
- Hiendleder, S.; Wirtz, M.; Mund, C.; Klempt, M.; Reichenbach, H.-D.; Stojkovic, M.; Weppert, M.; Wenigerkind, H.; Elmlinger, M.; Lyko, F.; et al. Tissue-specific effects of in vitro fertilization procedures on genomic cytosine methylation levels in overgrown and normal sized bovine fetuses. Biol. Reprod 2006, 75, 17–23. [Google Scholar]
- Curchoe, C.L.; Zhang, S.; Yang, L.; Page, R.; Tian, X.C. Hypomethylation trends in the intergenic region of the imprinted IGF2 and H19 genes in cloned cattle. Anim. Reprod. Sci 2009, 116, 213–225. [Google Scholar]
- Paoloni-Giacobino, A. Implications of reproductive technologies for birth and developmental outcomes: Imprinting defects and beyond. Expert Rev. Mol. Med 2006, 8, 1–14. [Google Scholar]
- Smith, L.; Suzuki, J., Jr; Goff, A.; Filion, F.; Therrien, J.; Murphy, B.; Kohan-Ghadr, H.; Lefebvre, R.; Brisville, A.; Buczinski, S. Epigenetic anomalies associated with prenatal survival and neonatal morbidity in cloned calves. Anim. Reprod. 2010, 7, 197–203. [Google Scholar]
- Heyman, Y.; Chavatte-Palmer, P.; LeBourhis, D.; Camous, S.; Vignon, X.; Renard, J.P. Frequency and occurrence of late-gestation losses from cattle cloned embryos. Biol. Reprod 2002, 66, 6–13. [Google Scholar]
- Cheng, X.; Hashimoto, H.; Horton, J.R.; Zhang, X. Mechanisms of DNA Methylation, Methyl-CpG Recognition, and Demethylation in Mammals. In Handbook of Epigenetics; Trygve, T., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 9–627. [Google Scholar]
- Dvir, A.; Conaway, J.W.; Conaway, R.C. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev 2001, 11, 209–214. [Google Scholar]
- Sandelin, A.; Carninci, P.; Lenhard, B.; Ponjavic, J.; Hayashizaki, Y.; Hume, D.A. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat. Rev. Genet 2007, 8, 424–436. [Google Scholar]
- Tran, K.; Gralla, J.D. Control of the timing of promoter escape and RNA catalysis by the transcription factor IIB fingertip. J. Biol. Chem 2008, 283, 15665–15671. [Google Scholar]
- Malik, S.; Roeder, R.G. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci 2005, 30, 256–263. [Google Scholar]
- Hoffmann, A.; Natoli, G.; Ghosh, G. Transcriptional regulation via the NF-κB signaling module. Oncogene 2006, 25, 6706–6716. [Google Scholar]
- Carrera, I.; Treisman, J.E. Message in a nucleus: Signaling to the transcriptional machinery. Curr. Opin. Genet. Dev 2008, 18, 397–403. [Google Scholar]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol 2010, 220, 126–139. [Google Scholar]
- Bonasio, R.; Tu, S.J.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar]
- Chotalia, M.; Smallwood, S.A.; Ruf, N.; Dawson, C.; Lucifero, D.; Frontera, M.; James, K.; Dean, W.; Kelsey, G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev 2009, 23, 105–117. [Google Scholar]
- Schmitt, S.; Prestel, M.; Paro, R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev 2005, 19, 697–708. [Google Scholar]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet 2009, 10, 295–304. [Google Scholar]
- Hanley, B.; Dijane, J.; Fewtrell, M.; Grynberg, A.; Hummel, S.; Junien, C.; Koletzko, B.; Lewis, S.; Renz, H.; Symonds, M.; et al. Metabolic imprinting, programming and epigenetics—A review of present priorities and future opportunities. Br. J. Nutr 2010, 104, S1–S25. [Google Scholar]
- Hore, T.A.; Rapkins, R.W.; Graves, J.A.M. Construction and evolution of imprinted loci in mammals. Trends Genet 2007, 23, 440–448. [Google Scholar]
- Yen, Z.C.; Meyer, I.M.; Karalic, S.; Brown, C.J. A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics 2007, 90, 453–463. [Google Scholar]
- Miranda, T.B.; Jones, P.A. DNA methylation: The nuts and bolts of repression. J. Cell. Physiol 2007, 213, 384–390. [Google Scholar]
- Lande-Diner, L.; Zhang, J.; Ben-Porath, I.; Amariglio, N.; Keshet, I.; Hecht, M.; Azuara, V.; Fisher, A.G.; Rechavi, G.; Cedar, H. Role of DNA methylation in stable gene repression. J. Biol. Chem 2007, 282, 12194–12200. [Google Scholar]
- Shi, L.J.; Wu, J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod. Biol. Endocrinol 2009, 7. [Google Scholar] [CrossRef]
- Wang, J.L.; Zhang, M.; Zhang, Y.; Kou, Z.H.; Han, Z.M.; Chen, D.Y.; Sun, Q.Y.; Gao, S.R. The histone demethylase JMJD2C is stage-specifically expressed in preimplantation mouse embryos and is required for embryonic development. Biol. Reprod 2010, 82, 105–111. [Google Scholar]
- Badr, H.; Bongioni, G.; Abdoon, A.S.S.; Kandil, O.; Puglisi, R. Gene expression in the in vitro-produced preimplantation bovine embryos. Zygote 2007, 15, 355–367. [Google Scholar]
- Hajkova, P. Epigenetic reprogramming—Taking a lesson from the embryo. Curr. Opin. Cell Biol 2010, 22, 342–350. [Google Scholar]
- Coan, P.M.; Burton, G.J.; Ferguson-Smith, A.C. Imprinted genes in the placenta—A review. Placenta 2005, 26, S10–S20. [Google Scholar]
- Fowden, A.L.; Coan, P.M.; Angiolini, E.; Burton, G.J.; Constancia, M. Imprinted genes and the epigenetic regulation of placental phenotype. Prog. Biophys. Mol. Biol 2011, 106, 281–288. [Google Scholar]
- Ng, H.K.; Novakovic, B.; Hiendleder, S.; Craig, J.M.; Roberts, C.T.; Saffery, R. Distinct patterns of gene-specific methylation in mammalian placentas: Implications for placental evolution and function. Placenta 2010, 31, 259–268. [Google Scholar]
- Schär, P.; Fritsch, O. DNA Repair and the Control of DNA Methylation. In Epigenetics and Disease; Gasser, S.M., Li, E., Eds.; Springer: Basel, Switzerland, 2011; Volume 67, pp. 51–68. [Google Scholar]
- Gronbaek, K.; Hother, C.; Jones, P.A. Epigenetic changes in cancer. APMIS 2007, 115, 1039–1059. [Google Scholar]
- Baumann, C.; Daly, C.M.; McDonnell, S.M.; Viveiros, M.M.; de la Fuente, R. Chromatin configuration and epigenetic landscape at the sex chromosome bivalent during equine spermatogenesis. Chromosoma 2011, 120, 227–244. [Google Scholar]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar]
- Lucia, P.; Fanti, L.; Negri, R.; Del Vescovo, V.; Fatica, A.; Pimpinelli, S. The Heterochromatin Protein 1 positively regulates euchromatic gene expression by RNA binding. Aviable online: http://hdl.handle.net/10101/npre.2008.2687.1 accessed on 27 July 2011.
- Girton, J.R.; Johansen, K.M. Chromatin Structure and the Regulation of Gene Expression: The Lessons of PEV in Drosophila. In Advances in Genetics; van Veronica, H., Robert, E.H., Eds.; Academic Press: San Diego, CA, USA, 2008; Volume 61, pp. 1–43. [Google Scholar]
- Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet 2002, 3, 662–673. [Google Scholar]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol 2009, 10, 697–708. [Google Scholar]
- Girod, P.-A.; Nguyen, D.-Q.; Calabrese, D.; Puttini, S.; Grandjean, M.; Martinet, D.; Regamey, A.; Saugy, D.; Beckmann, J.S.; Bucher, P.; et al. Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nat. Methods 2007, 4, 747–753. [Google Scholar]
- Shiota, K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet. Genome Res 2004, 105, 325–334. [Google Scholar]
- Kim, J.K.; Samaranayake, M.; Pradhan, S. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci 2009, 66, 596–612. [Google Scholar]
- Vaissiere, T.; Sawan, C.; Herceg, Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res 2008, 659, 40–48. [Google Scholar]
- Hanna, J.H.; Saha, K.; Jaenisch, R. Pluripotency and cellular reprogramming: Facts, hypotheses, unresolved issues. Cell 2010, 143, 508–525. [Google Scholar]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar]
- Weaver, J.R.; Susiarjo, M.; Bartolomei, M.S. Imprinting and epigenetic changes in the early embryo. Mamm. Genome 2009, 20, 532–543. [Google Scholar]
- Ooi, S.K.T.; O’Donnell, A.H.; Bestor, T.H. Mammalian cytosine methylation at a glance. J. Cell Sci 2009, 122, 2787–2791. [Google Scholar]
- Walsh, C.P.; Bestor, T.H. Cytosine methylation and mammalian development. Genes Dev 1999, 13, 26–34. [Google Scholar]
- Gopalakrishnan, S.; van Emburgh, B.O.; Robertson, K.D. DNA methylation in development and human disease. Mutat. Res 2008, 647, 30–38. [Google Scholar]
- Chang, H.; Zhang, T.; Zhang, Z.; Bao, R.; Fu, C.; Wang, Z.; Bao, Y.; Li, Y.; Wu, L.; Zheng, X.; et al. Tissue-specific distribution of aberrant DNA methylation associated with maternal low-folate status in human neural tube defects. J. Nutr. Biochem 2011, in press. [Google Scholar]
- Howlett, S.K.; Reik, W. Methylation levels of maternal and paternal genomes during preimplantation development. Development 1991, 113, 119–127. [Google Scholar]
- Monk, M.; Boubelik, M.; Lehnert, S. Temporal and regional changes in dna methylation in the embryonic, extraembryonic and germ-cell lineages during mouse embryo development. Development 1987, 99, 371–382. [Google Scholar]
- Gehring, M.; Reik, W.; Henikoff, S. DNA demethylation by DNA repair. Trends Genet 2009, 25, 82–90. [Google Scholar]
- Mayer, W.; Niveleau, A.; Walter, J.; Fundele, R.; Haaf, T. Embryogenesis: Demethylation of the zygotic paternal genome. Nature 2000, 403, 501–502. [Google Scholar]
- Oswald, J.; Engemann, S.; Lane, N.; Mayer, W.; Olek, A.; Fundele, R.; Dean, W.; Reik, W.; Walter, J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol 2000, 10, 475–478. [Google Scholar]
- Doherty, A.S.; Mann, M.R.W.; Tremblay, K.D.; Bartolomei, M.S.; Schultz, R.M. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod 2000, 62, 1526–1535. [Google Scholar]
- Mann, M.R.W.; Chung, Y.G.; Nolen, L.D.; Verona, R.I.; Latham, K.E.; Bartolomei, M.S. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod 2003, 69, 902–914. [Google Scholar]
- Kang, Y.K.; Lee, K.K.; Han, Y.M. Reprogramming DNA methylation in the preimplantation stage: Peeping with Dolly’s eyes. Curr. Opin. Cell Biol 2003, 15, 290–295. [Google Scholar]
- Jones, P.A.; Takai, D. The role of DNA methylation in mammalian epigenetics. Science 2001, 293, 1068–1070. [Google Scholar]
- Ferguson-Smith, A.C.; Surani, M.A. Imprinting and the epigenetic asymmetry between parental genomes. Science 2001, 293, 1086–1089. [Google Scholar]
- Mayer, W.; Niveleau, A.; Walter, J.; Fundele, R.; Haaf, T. Embryogenesis—Demethylation of the zygotic paternal genome. Nature 2000, 403, 501–502. [Google Scholar]
- Tremblay, K.D.; Saam, J.R.; Ingram, R.S.; Tilghman, S.M.; Bartolomei, M.S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet 1995, 9, 407–413. [Google Scholar]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar]
- Holmes, R.; Soloway, P.D. Regulation of imprinted DNA methylation. Cytogenet. Genome Res 2006, 113, 122–129. [Google Scholar]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol 2002, 241, 172–182. [Google Scholar]
- Gebert, C.; Wrenzycki, C.; Herrmann, D.; Groger, D.; Thiel, J.; Reinhardt, R.; Lehrach, H.; Hajkova, P.; Lucas-Hahn, A.; Carnwath, J.W.; et al. DNA methylation in the IGF2 intragenic DMR is re-established in a sex-specific manner in bovine blastocysts after somatic cloning. Genomics 2009, 94, 63–69. [Google Scholar]
- Durcova-Hills, G.; Hajkova, P.; Sullivan, S.; Barton, S.; Surani, M.A.; McLaren, A. Influence of sex chromosome constitution on the genomic imprinting of germ cells. Proc. Natl. Acad. Sci. USA 2006, 103, 11184–11188. [Google Scholar]
- Horsthemke, B. Genomic imprinting and imprinting defects. Med. Genet 2010, 22, 385–391. [Google Scholar]
- Hou, J.; Cui, X.H.; Lei, T.H.; Liu, L.; An, X.R.; Chen, Y.F. Aberrant DNA methylation patterns in cultured mouse embryos. Prog. Nat. Sci 2005, 15, 1079–1083. [Google Scholar]
- Beaujean, N.; Taylor, J.; Gardner, J.; Wilmut, I.; Meehan, R.; Young, L. Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biol. Reprod 2004, 71, 185–193. [Google Scholar]
- Wei, Y.; Zhu, J.; Huan, Y.; Liu, Z.; Yang, C.; Zhang, X.; Mu, Y.; Xia, P.; Liu, Z. Aberrant expression and methylation status of putatively imprinted genes in placenta of cloned piglets. Cell. Reprogram 2010, 12, 213–222. [Google Scholar]
- Bourque, D.K.; Avila, L.; Penaherrera, M.; von Dadelszen, P.; Robinson, W.P. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta 2010, 31, 197–202. [Google Scholar]
- Balassiano, K.; Lima, S.; Jenab, M.; Overvad, K.; Tjonneland, A.; Boutron-Ruault, M.C.; Clavel-Chapelon, F.; Canzian, F.; Kaaks, R.; Boeing, H.; et al. Aberrant DNA methylation of cancer-associated genes in gastric cancer in the european prospective investigation into cancer and nutrition (EPIC-EURGAST). Cancer Lett 2011, 311, 85–95. [Google Scholar]
- Chung, J.-H.; Lee, H.J.; Kim, B.-h.; Cho, N.-Y.; Kang, G.H. DNA methylation profile during multistage progression of pulmonary adenocarcinomas. Virchows Arch 2011, 459, 201–211. [Google Scholar]
- Estecio, M.R.H.; Issa, J.-P.J. Dissecting DNA hypermethylation in cancer. FEBS Lett 2011, 585, 2078–2086. [Google Scholar]
- Tada, Y.; Yokomizo, A.; Shiota, M.; Tsunoda, T.; Plass, C.; Naito, S. Aberrant DNA methylation of T-cell leukemia, homeobox 3 modulates cisplatin sensitivity in bladder cancer. Int. J. Oncol 2011, 39, 727–733. [Google Scholar]
- Shames, D.S.; Minna, J.D.; Gazdar, A.F. DNA methylation in health, disease, and cancer. Curr. Mol. Med 2007, 7, 85–102. [Google Scholar]
- Acevedo, L.G.; Sanz, A.; Jelinek, M.A. Novel DNA binding domain-based assays for detection of methylated and nonmethylated DNA. Epigenomics 2011, 3, 93–101. [Google Scholar]
- Laird, P.W. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet 2010, 11, 191–203. [Google Scholar]
- Harris, R.A.; Wang, T.; Coarfa, C.; Nagarajan, R.P.; Hong, C.B.; Downey, S.L.; Johnson, B.E.; Fouse, S.D.; Delaney, A.; Zhao, Y.J.; et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol 2010, 28, 1097–1105. [Google Scholar]
- Razin, A.; Kantor, B. DNA Methylation in Epigenetic Control of Gene Expression. In Epigenetics and Chromatin; Jeanteur, P., Ed.; Springer: Berlin, Germany, 2005; Volume 38, pp. 151–167. [Google Scholar]
- Singal, R.; Ginder, G.D. DNA methylation. Blood 1999, 93, 4059–4070. [Google Scholar]
- Bestor, T.; Laudano, A.; Mattaliano, R.; Ingram, V. Cloning and sequencing of a cDNA-encoding DNA methyltransferase of mouse cells: The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol 1988, 203, 971–983. [Google Scholar]
- Chen, T.P.; Li, E. Structure and Function of Eukaryotic DNA Methyltransferases. In Stem Cells in Development and Disease; Schatten, G.P., Ed.; Academic Press: San Diego, CA, USA, 2004; Volume 60, pp. 55–89. [Google Scholar]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet 2000, 9, 2395–2402. [Google Scholar]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G.P. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci 2010, 13, 423–430. [Google Scholar]
- Robert, M.F.; Morin, S.; Beaulieu, N.; Gauthier, F.; Chute, I.C.; Barsalou, A.; MacLeod, A.R. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet 2003, 33, 61–65. [Google Scholar]
- Chen, T.; Li, E. Establishment and maintenance of DNA methylation patterns in mammals. Curr. Top. Microbiol. Immunol 2006, 301, 179–201. [Google Scholar]
- Mortusewicz, O.; Schermelleh, L.; Walter, J.; Cardoso, M.C.; Leonhardt, H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. USA 2005, 102, 8905–8909. [Google Scholar]
- Grandjean, V.; Yaman, R.; Cuzin, F.; Rassoulzadegan, M. Inheritance of an epigenetic mark: The CpG DNA methyltransferase 1 is required for de novo establishment of a complex pattern of non-CpG methylation. PLoS One 2007, 2. [Google Scholar] [CrossRef]
- Schaefer, M.; Lyko, F. Solving the Dnmt2 enigma. Chromosoma 2010, 119, 35–40. [Google Scholar]
- Chedin, F. The DNMT3 family of mammalian de novo DNA methyltransferases. In Modifications of Nuclear DNA and Its Regulatory Proteins; Cheng, X.D., Blumenthal, R.M., Eds.; Academic Press: San Diego, CA, USA, 2011; Volume 101, pp. 255–285. [Google Scholar]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar]
- Kaneda, M.; Okano, M.; Hata, K.; Sado, T.; Tsujimoto, N.; Li, E.; Sasaki, H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004, 429, 900–903. [Google Scholar]
- Bourc’his, D.; Xu, G.L.; Lin, C.S.; Bollman, B.; Bestor, T.H. Dnmt3L and the establishment of maternal genomic imprints. Science 2001, 294, 2536–2539. [Google Scholar]
- Chedin, F.; Lieber, M.R.; Hsieh, C.L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 2002, 99, 16916–16921. [Google Scholar]
- Gowher, H.; Liebert, K.; Hermann, A.; Xu, G.L.; Jeltsch, A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem 2005, 280, 13341–13348. [Google Scholar]
- Webster, K.E.; O’Bryan, M.K.; Fletcher, S.; Crewther, P.E.; Aapola, U.; Craig, J.; Harrison, D.K.; Aung, H.; Phutikanit, N.; Lyle, R.; et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 4068–4073. [Google Scholar]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci 2006, 31, 89–97. [Google Scholar]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and function of mammalian DNA methyltransferases. Chembiochem 2011, 12, 206–222. [Google Scholar]
- Sakai, Y.; Suetake, I.; Shinozaki, F.; Yamashina, S.; Tajima, S. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr. Patterns 2004, 5, 231–237. [Google Scholar]
- Suetake, I.; Shinozaki, F.; Miyagawa, J.; Takeshima, H.; Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem 2004, 279, 27816–27823. [Google Scholar]
- Yamanaka, K.I.; Sakatani, M.; Kubota, K.; Balboula, A.Z.; Sawai, K.; Takahashi, M. Effects of downregulating DNA methyltransferase 1 transcript by RNA interference on DNA methylation status of the satellite I region and in vitro development of bovine somatic cell nuclear transfer embryos. J. Reprod. Dev 2011, 57, 393–402. [Google Scholar]
- Metivier, R.; Gallais, R.; Tiffoche, C.; Le Peron, C.; Jurkowska, R.Z.; Carmouche, R.P.; Ibberson, D.; Barath, P.; Demay, F.; Reid, G.; et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008, 452, 45–50. [Google Scholar]
- Ooi, S.L.; Henikoff, S. Germline histone dynamics and epigenetics. Curr. Opin. Cell Biol 2007, 19, 257–265. [Google Scholar]
- Gopalakrishnan, S.; Van Emburgh, B.O.; Shan, J.X.; Su, Z.; Fields, C.R.; Vieweg, J.; Hamazaki, T.; Schwartz, P.H.; Terada, N.; Robertson, K.D. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic DNA methylation patterns and displays altered DNA binding. Mol. Cancer Res 2009, 7, 1622–1634. [Google Scholar]
- Garzon, R.; Liu, S.J.; Fabbri, M.; Liu, Z.F.; Heaphy, C.E.A.; Callegari, E.; Schwind, S.; Pang, J.X.; Yu, J.H.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar]
- Hata, K.; Okano, M.; Lei, H.; Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002, 129, 1983–1993. [Google Scholar]
- Feltus, F.A.; Lee, E.K.; Costello, J.F.; Plass, C.; Vertino, P.M. Predicting aberrant CpG island methylation. Proc. Natl. Acad. Sci. USA 2003, 100, 12253–12258. [Google Scholar]
- Jair, K.W.; Bachman, K.E.; Suzuki, H.; Ting, A.H.; Rhee, I.; Yen, R.W.C.; Baylin, S.B.; Schuebel, K.E. De novo CpG island methylation in human cancer cells. Cancer Res 2006, 66, 682–692. [Google Scholar]
- Jia, D.; Jurkowska, R.Z.; Zhang, X.; Jeltsch, A.; Cheng, X.D. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 2007, 449, 248–251. [Google Scholar]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.Q.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.P.; Allis, C.D.; et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar]
- Ferguson-Smith, A.C.; Greally, J.M. Epigenetics: Perceptive enzymes. Nature 2007, 449, 148–149. [Google Scholar]
- Zhang, Y.Y.; Rohde, C.; Tierling, S.; Jurkowski, T.P.; Bock, C.; Santacruz, D.; Ragozin, S.; Reinhardt, R.; Groth, M.; Walter, J.; et al. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet 2009, 5. [Google Scholar] [CrossRef]
- Ooi, S.K.T.; Bestor, T.H. The colorful history of active DNA demethylation. Cell 2008, 133, 1145–1148. [Google Scholar]
- Hattori, N.; Imao, Y.; Nishino, K.; Ohgane, J.; Yagi, S.; Tanaka, S.; Shiota, K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells 2007, 12, 387–396. [Google Scholar]
- Simonsson, S.; Gurdon, J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat. Cell Biol 2004, 6, 984–990. [Google Scholar]
- La Salle, S.; Mertineit, C.; Taketo, T.; Moens, P.B.; Bestor, T.H.; Trasler, J.M. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol 2004, 268, 403–415. [Google Scholar]
- Lees-Murdock, D.J.; Shovlin, T.C.; Gardiner, T.; De Felici, M.; Walsh, C.P. DNA methyltransferase expression in the mouse germ line during periods of de novo methylation. Dev. Dyn 2005, 232, 992–1002. [Google Scholar]
- Davis, T.L.; Yang, G.J.; McCarrey, J.R.; Bartolomei, M.S. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet 2000, 9, 2885–2894. [Google Scholar]
- Fedoriw, A.M.; Stein, P.; Svoboda, P.; Schultz, R.M.; Bartolomei, M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 2004, 303, 238–240. [Google Scholar]
- Jelinic, P.; Stehle, J.C.; Shaw, P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol 2006, 4, e355. [Google Scholar]
- Verona, R.I.; Mann, M.R.W.; Bartolomei, M.S. Genomic imprinting: Intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol 2003, 19, 237–259. [Google Scholar]
- Fitzpatrick, G.V.; Soloway, P.D.; Higgins, M.J. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet 2002, 32, 426–431. [Google Scholar]
- Mancini-DiNardo, D.; Steele, S.J.S.; Ingram, R.S.; Tilghman, S.M. A differentially methylated region within the gene KCNQ1 functions as an imprinted promoter and silencer. Hum. Mol. Genet 2003, 12, 283–294. [Google Scholar]
- Thorvaldsen, J.L.; Duran, K.L.; Bartolomei, M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and IGF2. Genes Dev 1998, 12, 3693–3702. [Google Scholar]
- Zhang, Y.J.; Qu, L.H. Non-coding RNAs and the acquisition of genomic imprinting in mammals. Sci. China C Life Sci 2009, 52, 195–204. [Google Scholar]
- Peters, J.; Robson, J.E. Imprinted noncoding RNAs. Mamm. Genome 2008, 19, 493–502. [Google Scholar]
- Latos, P.A.; Barlow, D.P. Regulation of imprinted expression by macro non-coding RNAs. RNA Biol 2009, 6, 100–106. [Google Scholar]
- Bourc’his, D.; Voinnet, O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 2010, 330, 617–622. [Google Scholar]
- Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009, 457, 413–420. [Google Scholar]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar]
- Koerner, M.V.; Pauler, F.M.; Huang, R.; Barlow, D.P. The function of non-coding RNAs in genomic imprinting. Development 2009, 136, 1771–1783. [Google Scholar]
- Erhard, F.; Zimmer, R. Classification of ncRNAs using position and size information in deep sequencing data. Bioinformatics 2010, 26, i426–i432. [Google Scholar]
- Childs, L.; Nikoloski, Z.; May, P.; Walther, D. Identification and classification of ncRNA molecules using graph properties. Nucleic Acids Res 2009, 37. [Google Scholar] [CrossRef]
- Wutz, A.; Gribnau, J. X inactivation Xplained. Curr. Opin. Genet. Dev 2007, 17, 387–393. [Google Scholar]
- Seitz, H.; Royo, H.; Lin, S.P.; Youngson, N.; Ferguson-Smith, A.C.; Cavaille, J. Imprinted small RNA genes. Biol. Chem 2004, 385, 905–911. [Google Scholar]
- Royo, H.; Bortolin, M.L.; Seitz, H.; Cavaille, J. Small non-coding RNAs and genomic imprinting. Cytogenet. Genome Res 2006, 113, 99–108. [Google Scholar]
- Wang, Y.; Medvid, R.; Melton, C.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet 2007, 39, 380–385. [Google Scholar]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005, 19, 489–501. [Google Scholar]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol 2008, 9, 219–230. [Google Scholar]
- Santoro, F.; Barlow, D.P. Developmental control of imprinted expression by macro non-coding RNAs. Semin. Cell Dev. Biol 2011, 22, 328–335. [Google Scholar]
- Martello, G.; Zacchigna, L.; Inui, M.; Montagner, M.; Adorno, M.; Mamidi, A.; Morsut, L.; Soligo, S.; Tran, U.; Dupont, S.; et al. MicroRNA control of nodal signalling. Nature 2007, 449, 183–188. [Google Scholar]
- Kwon, C.; Han, Z.; Olson, E.N.; Srivastava, D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates notch signaling. Proc. Natl. Acad. Sci. USA 2005, 102, 18986–18991. [Google Scholar]
- Deng, Z.; Chen, J.-F.; Wang, D.-Z. Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskelet. Disord 2011, 12. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004, 5, R13–R23. [Google Scholar]
- Vo, N.K.; Cambronne, X.A.; Goodman, R.H. MicroRNA pathways in neural development and plasticity. Curr. Opin. Neurobiol 2010, 20, 457–465. [Google Scholar]
- Hudson, Q.J.; Kulinski, T.M.; Huetter, S.P.; Barlow, D.P. Genomic imprinting mechanisms in embryonic and extraembryonic mouse tissues. Heredity 2010, 105, 45–56. [Google Scholar]
- Lewis, A.; Mitsuyaj, K.; Constancia, M.; Reik, W. Tandem repeat hypothesis in imprinting: Deletion of a conserved direct repeat element upstream of H19 has no effect on imprinting in the IGF2-H19 region. Mol. Cell. Biol 2004, 24, 5650–5656. [Google Scholar]
- Umlauf, D.; Goto, Y.; Cao, R.; Cerqueira, F.; Wagschal, A.; Zhang, Y.; Feil, R. Imprinting along the KCNQ1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet 2004, 36, 1296–1300. [Google Scholar]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-DiNardo, D.; Kanduri, C. Antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar]
- Terranova, R.; Yokobayashi, S.; Stadler, M.B.; Otte, A.P.; van Lohuizen, M.; Orkin, S.H.; Peters, A.H.F.M. Polycomb group proteins EZH2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell 2008, 15, 668–679. [Google Scholar]
- Carmell, M.A.; Hannon, G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol 2004, 11, 214–218. [Google Scholar]
- Chu, C.-Y.; Rana, T.M. Small RNAs: Regulators and guardians of the genome. J. Cell. Physiol 2007, 213, 412–419. [Google Scholar]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet 2008, 9, 102–114. [Google Scholar]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol 2009, 10, 126–139. [Google Scholar]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar]
- Klattenhoff, C.; Theurkauf, W. Biogenesis and germline functions of piRNAs. Development 2008, 135, 3–9. [Google Scholar]
- Szakmary, A.; Cox, D.N.; Wang, Z.; Lin, H.F. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol 2005, 15, 171–178. [Google Scholar]
- Efroni, S.; Duttagupta, R.; Cheng, J.; Dehghani, H.; Hoeppner, D.J.; Dash, C.; Bazett-Jones, D.P.; Le Grice, S.; McKay, R.D.G.; Buetow, K.H.; et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008, 2, 437–447. [Google Scholar]
- Kimura, H.; Tada, M.; Nakatsuji, N.; Tada, T. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol. Cell. Biol 2004, 24, 5710–5720. [Google Scholar]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar]
- Kim, J.M.; Liu, H.L.; Tazaki, M.; Nagata, M.; Aoki, F. Changes in histone acetylation during mouse oocyte meiosis. J. Cell Biol 2003, 162, 37–46. [Google Scholar]
- Kimmins, S.; Sassone-Corsi, P. Chromatin remodelling and epigenetic features of germ cells. Nature 2005, 434, 583–589. [Google Scholar]
- Yamanaka, K.; Sugimura, S.; Wakai, T.; Kawahara, M.; Sato, E. Acetylation level of histone H3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J. Reprod. Dev 2009, 55, 638–644. [Google Scholar]
- Zhang, Y.H.; Li, J.; Villemoes, K.; Pedersen, A.M.; Purup, S.; Vajta, G. An epigenetic modifier results in improved in vitro blastocyst production after somatic cell nuclear transfer. Cloning Stem Cells 2007, 9, 357–363. [Google Scholar]
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigo, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.P.; Snyder, M.; Dermitzakis, E.T.; Thurman, R.E.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar]
- Tamaru, H.; Selker, E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 2001, 414, 277–283. [Google Scholar]
- Jackson, J.P.; Lindroth, A.M.; Cao, X.F.; Jacobsen, S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 2002, 416, 556–560. [Google Scholar]
- Fuks, F. DNA methylation and histone modifications: Teaming up to silence genes. Curr. Opin. Genet. Dev 2005, 15, 490–495. [Google Scholar]
- Francis, N.J.; Follmer, N.E.; Simon, M.D.; Aghia, G.; Butler, J.D. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell 2009, 137, 110–122. [Google Scholar]
- Blobel, G.A.; Kadauke, S.; Wang, E.; Lau, A.W.; Zuber, J.; Chou, M.M.; Vakoc, C.R. A reconfigured pattern of Mll occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 2009, 36, 970–983. [Google Scholar]
- Hahn, M.A.; Wu, X.W.; Li, A.X.; Hahn, T.; Pfeifer, G.P. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Bartke, T.; Vermeulen, M.; Xhemalce, B.; Robson, S.C.; Mann, M.; Kouzarides, T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010, 143, 470–484. [Google Scholar]
- Lindroth, A.M.; Park, Y.J.; McLean, C.M.; Dokshin, G.A.; Persson, J.M.; Herman, H.; Pasini, D.; Miro, X.; Donohoe, M.E.; Lee, J.T.; et al. Antagonism between DNA and H3K27 methylation at the imprinted RASGRF1 locus. PLoS Genet 2008, 4. [Google Scholar] [CrossRef]
- Myant, K.; Termanis, A.; Sundaram, A.Y.M.; Boe, T.; Li, C.; Merusi, C.; Burrage, J.; de Las Heras, J.I.; Stancheva, I. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res 2011, 21, 83–94. [Google Scholar]
- Fournier, C.; Goto, Y.J.; Ballestar, E.; Delaval, K.; Hever, A.M.; Esteller, M.; Feil, R. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J 2002, 21, 6560–6570. [Google Scholar]
- Rougeulle, C.; Navarro, P.; Avner, P. Promoter-restricted H3 Lys 4 di-methylation is an epigenetic mark for monoallelic expression. Hum. Mol. Genet 2003, 12, 3343–3348. [Google Scholar]
- Appanah, R.; Dickerson, D.R.; Goyal, P.; Groudine, M.; Lorincz, M.C. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet 2007, 3, 241–253. [Google Scholar]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Paabo, S.; Rebhan, M.; Schubeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet 2007, 39, 457–466. [Google Scholar]
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The Polycomb-group gene EZH2 is required for early mouse development. Mol. Cell. Biol 2001, 21, 4330–4336. [Google Scholar]
- Tachibana, M.; Sugimoto, K.; Nozaki, M.; Ueda, J.; Ohta, T.; Ohki, M.; Fukuda, M.; Takeda, N.; Niida, H.; Kato, H.; et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 2002, 16, 1779–1791. [Google Scholar]
- Lagger, G.; O’Carroll, D.; Rembold, M.; Khier, H.; Tischler, J.; Weitzer, G.; Schuettengruber, B.; Hauser, C.; Brunmeir, R.; Jenuwein, T.; et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 2002, 21, 2672–2681. [Google Scholar]
- Tilghman, S.M. The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell 1999, 96, 185–193. [Google Scholar]
- Wagschal, A.; Feil, R. Genomic imprinting in the placenta. Cytogenet. Genome Res 2006, 113, 90–98. [Google Scholar]
- Davies, W.; Smith, R.J.; Kelsey, G.; Wilkinson, L.S. Expression patterns of the novel imprinted genes Nap1l5 and Peg13 and their non-imprinted host genes in the adult mouse brain. Gene Expr. Patterns 2004, 4, 741–747. [Google Scholar]
- Smith, R.J.; Dean, W.; Konfortova, G.; Kelsey, G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res 2003, 13, 558–569. [Google Scholar]
- Kagitani, F.; Kuroiwa, Y.; Wakana, S.; Shiroishi, T.; Miyoshi, N.; Kobayashi, S.; Nishida, M.; Kohda, T.; KanekoIshino, T.; Ishino, F. Peg5/Neuronatin is an imprinted gene located on sub-distal chromosome 2 in the mouse. Nucleic Acids Res 1997, 25, 3428–3432. [Google Scholar]
- Kikyo, N.; Williamson, C.M.; John, R.M.; Barton, S.C.; Beechey, C.V.; Ball, S.T.; Cattanach, B.M.; Surani, M.A.; Peters, J. Genetic and functional analysis of neuronatin in mice with maternal or paternal duplication of distal Chr 2. Dev. Biol 1997, 190, 66–77. [Google Scholar]
- Choi, J.D.; Underkoffler, L.A.; Wood, A.J.; Collins, J.N.; Williams, P.T.; Golden, J.A.; Schuster, E.F.; Loomes, K.M.; Oakey, R.J. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol. Cell. Biol 2005, 25, 5514–5522. [Google Scholar]
- Peters, J.; Beechey, C. Identification and characterisation of imprinted genes in the mouse. Brief. Funct. Genomics Proteomics 2004, 2, 320–333. [Google Scholar]
- Branco, M.R.; Oda, M.; Reik, W. Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev 2008, 22, 1567–1571. [Google Scholar]
- Ikegami, K.; Ohgane, J.; Tanaka, S.; Yagi, S.; Shiota, K. Interplay between DNA methylation, histone modification and chromatin remodeling in stem cells and during development. Int. J. Dev. Biol 2009, 53, 203–214. [Google Scholar]
- Royo, H.; Cavaille, J. Non-coding RNAs in imprinted gene clusters. Biol. Cell 2008, 100, 149–166. [Google Scholar]
- Wutz, A.; Smrzka, O.W.; Schweifer, N.; Schellander, K.; Wagner, E.F.; Barlow, D.P. Imprinted expression of the IGF2r gene depends on an intronic CpG island. Nature 1997, 389, 745–749. [Google Scholar]
- Birger, Y.; Shemer, R.; Perk, J.; Razin, A. The imprinting box of the mouse IGF2r gene. Nature 1999, 397, 84–88. [Google Scholar]
- Kantor, B.; Makedonski, K.; Green-Finberg, Y.; Shemer, R.; Razin, A. Control elements within the PWS/AS imprinting box and their function in the imprinting process. Hum. Mol. Genet 2004, 13, 751–762. [Google Scholar]
- Ideraabdullah, F.Y.; Abramowitz, L.K.; Thorvaldsen, J.L.; Krapp, C.; Wen, S.C.; Engel, N.; Bartolomei, M.S. Novel cis-regulatory function in ICR-mediated imprinted repression of H19. Dev. Biol 2011, 355, 349–357. [Google Scholar]
- Edwards, C.A.; Ferguson-Smith, A.C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol 2007, 19, 281–289. [Google Scholar]
- Brandeis, M.; Kafri, T.; Ariel, M.; Chaillet, J.R.; McCarrey, J.; Razin, A.; Cedar, H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J 1993, 12, 3669–3677. [Google Scholar]
- Moore, T.; Constancia, M.; Zubair, M.; Bailleul, B.; Feil, R.; Sasaki, H.; Reik, W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse IGF2. Proc. Natl. Acad. Sci. USA 1997, 94, 12509–12514. [Google Scholar]
- Feil, R.; Walter, J.; Allen, N.D.; Reik, W. Developmental control of allelic methylation in the imprinted mouse IGF2 and H19 genes. Development 1994, 120, 2933–2943. [Google Scholar]
- Kacem, S.; Feil, R. Chromatin mechanisms in genomic imprinting. Mamm. Genome 2009, 20, 544–556. [Google Scholar]
- Hirasawa, R.; Chiba, H.; Kaneda, M.; Tajima, S.; Li, E.; Jaenisch, R.; Sasaki, H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 2008, 22, 1607–1616. [Google Scholar]
- Yang, L.; Chavatte-Palmer, P.; Kubota, C.; O’Neill, M.; Hoagland, T.; Renard, J.P.; Taneja, M.; Yang, X.Z.; Tian, X.C. Expression of imprinted genes is aberrant in deceased newborn cloned calves and relatively normal in surviving adult clones. Mol. Reprod. Dev 2005, 71, 431–438. [Google Scholar]
- Zhang, S.Q.; Kubota, C.; Yang, L.; Zhang, Y.Q.; Page, R.; O’Neill, M.; Yang, X.Z.; Tian, X.C. Genomic imprinting of H19 in naturally reproduced and cloned cattle. Biol. Reprod 2004, 71, 1540–1544. [Google Scholar]
- Sato, A.; Otsu, E.; Negishi, H.; Utsunomiya, T.; Arima, T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum. Reprod 2007, 22, 26–35. [Google Scholar]
- Market-Velker, B.A.; Zhang, L.Y.; Magri, L.S.; Bonvissuto, A.C.; Mann, M.R.W. Dual effects of superovulation: Loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum. Mol. Genet 2010, 19, 36–51. [Google Scholar]
- Park, C.H.; Kim, H.S.; Lee, S.G.; Lee, C.K. Methylation status of differentially methylated regions at IGF2/H19 locus in porcine gametes and preimplantation embryos. Genomics 2009, 93, 179–186. [Google Scholar]
- Suzuki, J.; Therrien, J.; Filion, F.; Lefebvre, R.; Goff, A.K.; Perecin, F.; Meirelles, F.V.; Smith, L.C. Loss of methylation at H19 DMD is associated with biallelic expression and reduced development in cattle derived by somatic cell nuclear transfer. Biol. Reprod 2011, 84, 947–956. [Google Scholar]
- Morgan, H.D.; Santos, F.; Green, K.; Dean, W.; Reik, W. Epigenetic reprogramming in mammals. Hum. Mol. Genet 2005, 14, R47–R58. [Google Scholar]
- Sassone-Corsi, P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 2002, 296, 2176–2178. [Google Scholar]
- Pathak, S.; Kedia-Mokashi, N.; Saxena, M.; D’Souza, R.; Maitra, A.; Parte, P.; Gill-Sharma, M.; Balasinor, N. Effect of tamoxifen treatment on global and insulin-like growth factor 2-H19 locus-specific DNA methylation in rat spermatozoa and its association with embryo loss. Fertil. Steril 2009, 91, 2253–2263. [Google Scholar]
- Delaval, K.; Feil, R. Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev 2004, 14, 188–195. [Google Scholar]
- McLay, D.W.; Clarke, H.J. Remodelling the paternal chromatin at fertilization in mammals. Reproduction 2003, 125, 625–633. [Google Scholar]
- Labosky, P.A.; Barlow, D.P.; Hogan, B.L.M. Mouse embryonic germ (EG) cell-lines: Transmission through the germline and differences in the methylation imprint of insulin-like growth-factor 2 receptor (IGF2r) gene compared with embryonic stem (ES) cell-lines. Development 1994, 120, 3197–3204. [Google Scholar]
- Tada, T.; Tada, M.; Hilton, K.; Barton, S.C.; Sado, T.; Takagi, N.; Surani, M.A. Epigenotype switching of imprintable loci in embryonic germ cells. Dev. Genes Evol 1998, 207, 551–561. [Google Scholar]
- Durcova-Hills, G.; Burgoyne, P.; McLaren, A. Analysis of sex differences in EGC imprinting. Dev. Biol 2004, 268, 105–110. [Google Scholar]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 2008, 31, 785–799. [Google Scholar]
- Carmell, M.A.; Girard, A.; van de Kant, H.J.G.; Bourc’his, D.; Bestor, T.H.; de Rooij, D.G.; Hannon, G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 2007, 12, 503–514. [Google Scholar]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W.; et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 2008, 22, 908–917. [Google Scholar]
- Nicholas, C.R.; Chavez, S.L.; Baker, V.L.; Pera, R.A.R. Instructing an embryonic stem cell-derived oocyte fate: Lessons from endogenous oogenesis. Endocr. Rev 2009, 30, 264–283. [Google Scholar]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, 539–543. [Google Scholar]
- Yoon, B.J.; Herman, H.; Sikora, A.; Smith, L.T.; Plass, C.; Soloway, P.D. Regulation of DNA methylation of RASGRF1. Nat. Genet 2002, 30, 92–96. [Google Scholar]
- Holmes, R.; Chang, Y.J.; Soloway, P.D. Timing and sequence requirements defined for embryonic maintenance of imprinted DNA methylation at RASGRF1. Mol. Cell. Biol 2006, 26, 9564–9570. [Google Scholar]
- Constancia, M.; Hemberger, M.; Hughes, J.; Dean, W.; Ferguson-Smith, A.; Fundele, R.; Stewart, F.; Kelsey, G.; Fowden, A.; Sibley, C.; et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 2002, 417, 945–948. [Google Scholar]
- Kalscheuer, V.M.; Mariman, E.C.; Schepens, M.T.; Rehder, H.; Ropers, H.H. The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nat. Genet 1993, 5, 74–78. [Google Scholar]
- Gardner, D.K.; Larman, M.G.; Thouas, G.A. Sex-related physiology of the preimplantation embryo. Mol. Hum. Reprod 2010, 16, 539–547. [Google Scholar]
- Biliya, S.; Bulla, L.A. Genomic imprinting: The influence of differential methylation in the two sexes. Exp. Biol. Med 2010, 235, 139–147. [Google Scholar]
- Hajkova, P.; Erhardt, S.; Lane, N.; Haaf, T.; El-Maarri, O.; Reik, W.; Walter, J.; Surani, M.A. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev 2002, 117, 15–23. [Google Scholar]
- Yamazaki, Y.; Mann, M.R.W.; Lee, S.S.; Marh, J.; McCarrey, J.R.; Yanagimachi, R.; Bartolomei, M.S. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc. Natl. Acad. Sci. USA 2003, 100, 12207–12212. [Google Scholar]
- Hajkova, P.; Ancelin, K.; Waldmann, T.; Lacoste, N.; Lange, U.C.; Cesari, F.; Lee, C.; Almouzni, G.; Schneider, R.; Surani, M.A. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 2008, 452, 877–881. [Google Scholar]
- Hazzouri, M.; Pivot-Pajot, C.; Faure, A.K.; Usson, Y.; Pelletier, R.; Sele, B.; Khochbin, S.; Rousseaux, S. Regulated hyperacetylation of core histones during mouse spermatogenesis: Involvement of histone-deacetylases. Eur. J. Cell Biol 2000, 79, 950–960. [Google Scholar]
- Szabo, P.E.; Mann, J.R. Biallelic expression of imprinted genes in the mouse germ-line: Implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev 1995, 9, 1857–1868. [Google Scholar]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar]
- Ku, M.; Koche, R.P.; Rheinbay, E.; Mendenhall, E.M.; Endoh, M.; Mikkelsen, T.S.; Presser, A.; Nusbaum, C.; Xie, X.H.; Chi, A.S.; et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008, 4. [Google Scholar] [CrossRef]
- Kashyap, V.; Rezende, N.C.; Scotland, K.B.; Shaffer, S.M.; Persson, J.L.; Gudas, L.J.; Mongan, N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the Nanog, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev 2009, 18, 1093–1108. [Google Scholar]
- Atkinson, S.; Armstrong, L. Epigenetics in embryonic stem cells: Regulation of pluripotency and differentiation. Cell Tissue Res 2008, 331, 23–29. [Google Scholar]
- Pasini, D.; Bracken, A.P.; Hansen, J.B.; Capillo, M.; Helin, K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol 2007, 27, 3769–3779. [Google Scholar]
- Herranz, N.; Pasini, D.; Diaz, V.M.; Franci, C.; Gutierrez, A.; Dave, N.; Escriva, M.; Hernandez-Munoz, I.; di Croce, L.; Helin, K.; et al. Polycomb complex 2 is required for E-cadherin repression by the snail1 transcription factor. Mol. Cell. Biol 2008, 28, 4772–4781. [Google Scholar]
- Yuzyuk, T.; Fakhouri, T.H.I.; Kiefer, J.; Mango, S.E. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev. Cell 2009, 16, 699–710. [Google Scholar]
- Iwahashi, K.; Yoshioka, H.; Low, E.W.; McCarrey, J.R.; Yanagimachi, R.; Yamazaki, Y. Autonomous regulation of sex-specific developmental programming in mouse fetal germ cells. Biol. Reprod 2007, 77, 697–706. [Google Scholar]
- Feil, R.; Berger, F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 2007, 23, 192–199. [Google Scholar]
- Wagschal, A.; Sutherland, H.G.; Woodfine, K.; Henckel, A.; Chebli, K.; Schulz, R.; Oakey, R.J.; Bickmore, W.A.; Feil, R. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell. Biol 2008, 28, 1104–1113. [Google Scholar]
- Ono, R.; Kobayashi, S.; Wagatsuma, H.; Aisaka, K.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 2001, 73, 232–237. [Google Scholar]
- Sekita, Y.; Wagatsuma, H.; Nakamura, K.; Ono, R.; Kagami, M.; Wakisaka, N.; Hino, T.; Suzuki-Migishima, R.; Kohda, T.; Ogura, A.; et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet 2008, 40, 243–248. [Google Scholar]
- Haycock, P.C.; Ramsay, M. Exposure of mouse embryos to ethanol during preimplantation development: Effect on dna methylation in the H19 imprinting control region. Biol. Reprod 2009, 81, 618–627. [Google Scholar]
- Su, J.M.; Xu, W.B.; Li, Y.Y.; Wang, L.J.; Wang, Y.S.; Zhang, Y. The methylation status of PEG10 in placentas of cloned transgenic calves. Yi Chuan 2011, 33, 533–538. [Google Scholar]
- Vire, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; van Eynde, A.; Bernard, D.; Vanderwinden, J.M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006, 439, 871–874. [Google Scholar]
- Pasini, D.; Bracken, A.P.; Agger, K.; Christensen, J.; Hansen, K.; Cloos, P.A.C.; Helin, K. Regulation of Stem Cell Differentiation by Histone Methyltransferases and Demethylases. Cold Spring Harb. Symp. Quant. Biol 2008, 73, 253–263. [Google Scholar]
- Schuettengruber, B.; Chourrout, D.; Vervoort, M.; Leblanc, B.; Cavalli, G. Genome regulation by polycomb and trithorax proteins. Cell 2007, 128, 735–745. [Google Scholar]
- Bantignies, F.; Cavalli, G. Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol 2006, 18, 275–283. [Google Scholar]
- Ross, P.J.; Ragina, N.P.; Rodriguez, R.M.; Iager, A.E.; Siripattarapravat, K.; Lopez-Corrales, N.; Cibelli, J.B. Polycomb gene expression and histone H3 lysine 27 trimethylation changes during bovine preimplantation development. Reproduction 2008, 136, 777–785. [Google Scholar]
- Dunn, K.L.; Davie, J.R. The many roles of the transcriptional regulator CTCF. Biochem. Cell Biol 2003, 81, 161–167. [Google Scholar]
- Filippova, G.N.; Fagerlie, S.; Klenova, E.M.; Myers, C.; Dehner, Y.; Goodwin, G.; Neiman, P.E.; Collins, S.J.; Lobanenkov, V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-Myc oncogenes. Mol. Cell. Biol 1996, 16, 2802–2813. [Google Scholar]
- Soshnikova, N.; Montavon, T.; Leleu, M.; Galjart, N.; Duboule, D. Functional analysis of CTCF during mammalian limb development. Dev. Cell 2010, 19, 819–830. [Google Scholar]
- Essien, K.; Vigneau, S.; Apreleva, S.; Singh, L.N.; Bartolomei, M.S.; Hannenhalli, S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol 2009, 10. [Google Scholar] [CrossRef]
- Vostrov, A.A.; Quitschke, W.W. The zinc finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter. J. Biol. Chem 1997, 272, 33353–33359. [Google Scholar]
- Cuddapah, S.; Jothi, R.; Schones, D.E.; Roh, T.Y.; Cui, K.R.; Zhao, K.J. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 2009, 19, 24–32. [Google Scholar]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar]
- Han, L.; Lee, D.H.; Szabo, P.E. CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/IGF2 imprinted region. Mol. Cell. Biol 2008, 28, 1124–1135. [Google Scholar]
- Kim, T.H.; Abdullaev, Z.K.; Smith, A.D.; Ching, K.A.; Loukinov, D.I.; Green, R.D.; Zhang, M.Q.; Lobanenkov, V.V.; Ren, B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007, 128, 1231–1245. [Google Scholar]
- Ideraabdullah, F.Y.; Vigneau, S.; Bartolomei, M.S. Genomic imprinting mechanisms in mammals. Mutat. Res 2008, 647, 77–85. [Google Scholar]
- Matsuzaki, H.; Okamura, E.; Fukamizu, A.; Tanimoto, K. CTCF binding is not the epigenetic mark that establishes post-fertilization methylation imprinting in the transgenic H19 ICR. Hum. Mol. Genet 2010, 19, 1190–1198. [Google Scholar]
- Esteve, P.-O.; Chin, H.G.; Smallwood, A.; Feehery, G.R.; Gangisetty, O.; Karpf, A.R.; Carey, M.F.; Pradhan, S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 2006, 20, 3089–3103. [Google Scholar]
- Robertson, A.K.; Geiman, T.M.; Sankpal, U.T.; Hager, G.L.; Robertson, K.D. Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem. Biophys. Res. Commun 2004, 322, 110–118. [Google Scholar]
| DNMT # types [100] | Functions |
|---|---|
| DNMT1 | Maintaining methylation pattern [21,101,102] |
| Essential for chromosome replication and repair [21,103,104] | |
| Essential for de novo methylation [105] | |
| DNMT2 | Effective in DNA and RNA methylation (for review see [106]) |
| DNMT3a | Establishment of de novo methylation pattern [107,108] |
| especially during gametogenesis [109] | |
| Maintaining methylation pattern [101] | |
| DNMT3b | Establishment of de novo methylation [107,108] |
| DNMT3L | Essential for de novo methylation [110] |
| Enhance de novo methylation activity of DNMT3a [111] and DNMT3b [112] | |
| Establishment of de novo methylation pattern especially during gametogenesis [113] | |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Golbabapour, S.; Abdulla, M.A.; Hajrezaei, M. A Concise Review on Epigenetic Regulation: Insight into Molecular Mechanisms. Int. J. Mol. Sci. 2011, 12, 8661-8694. https://doi.org/10.3390/ijms12128661
Golbabapour S, Abdulla MA, Hajrezaei M. A Concise Review on Epigenetic Regulation: Insight into Molecular Mechanisms. International Journal of Molecular Sciences. 2011; 12(12):8661-8694. https://doi.org/10.3390/ijms12128661
Chicago/Turabian StyleGolbabapour, Shahram, Mahmood Ameen Abdulla, and Maryam Hajrezaei. 2011. "A Concise Review on Epigenetic Regulation: Insight into Molecular Mechanisms" International Journal of Molecular Sciences 12, no. 12: 8661-8694. https://doi.org/10.3390/ijms12128661




