Self-Assembly in the Ferritin Nano-Cage Protein Superfamily
Abstract
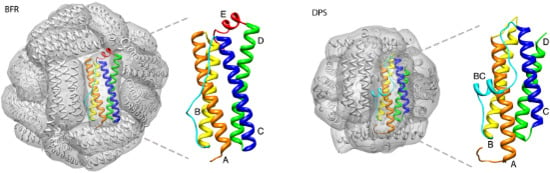
:1. Introduction
2. Maxi-Ferritin Self-Assembly
2.1. Proposed Pathways of Maxi-Ferritin Assembly
2.2. Mutation Related Studies on the Effect of Maxi-Ferritins Self-Assembly
3. Mini-Ferritin Self-Assembly
3.1. Proposed Pathways of Mini-Ferritin Assembly
3.2. Mutation Related Studies on the Effect of Mini-Ferritins Self-Assembly
3.3. pH Effects on the Self-Assembly of Mini-Ferritin
4. Conclusions
Acknowledgments
References
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar]
- Peczuh, M; Orner, B; Hamilton, A. In recognition of proteins. Chem. Br 2000, 36, 43–45. [Google Scholar]
- Ghadiri, MR; Granja, JR; Milligan, RA; Mcree, DE; Khazanovich, N. Self-Assembling organic nanotubes based on a cyclic peptide architecture. Nature 1993, 366, 324–327. [Google Scholar]
- Gauthier, D; Baillargeon, P; Drouin, M; Dory, YL. Self-Assembly of cyclic peptides into nanotubes and then into highly anisotropic crystalline materials this work was supported by FCAR quebec. Angew. Chem. Int. Ed. Engl 2001, 40, 4635–4638. [Google Scholar]
- Sanchez-Quesada, J; Ghadiri, MR; Bayley, H; Braha, O. Cyclic peptides as molecular adapters for a pore-forming protein. J. Am. Chem. Soc 2000, 122, 11757–11766. [Google Scholar]
- Bong, DT; Ghadiri, MR. Self-assembling cyclic peptide cylinders as nuclei for crystal engineering. Angew. Chem. Int. Ed. Engl 2001, 40, 2163–2166. [Google Scholar]
- Dotan, N; Arad, D; Frolow, F; Freeman, A. Self-assembly of a tetrahedral lectin into predesigned diamondlike protein crystals. Angew. Chem. Int. Ed. Engl 1999, 38, 2363–2366. [Google Scholar]
- Padilla, JE; Colovos, C; Yeates, TO. Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl. Acad. Sci. USA 2001, 98, 2217–2221. [Google Scholar]
- Zlotnick, A. To build a virus capsid: an equilibrium-model of the self-assembly of polyhedral protein complexes. J. Mol. Biol 1994, 241, 59–67. [Google Scholar]
- Fraenkelconrat, H; Williams, RC. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. USA 1955, 41, 690–698. [Google Scholar]
- Villar, G; Wilber, AW; Williamson, AJ; Thiara, P; Doye, JPK; Louis, AA; Jochum, MN; Lewis, ACF; Levy, ED. Self-Assembly and evolution of homomeric protein complexes. Phys. Rev. Lett 2009, 102, 118106. [Google Scholar]
- Crick, FH; Watson, JD. Structure of small viruses. Nature 1956, 177, 473–475. [Google Scholar]
- Berger, W; Steiner, E; Grusch, M; Elbling, L; Micksche, M. Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell. Mol. Life Sci 2009, 66, 43–61. [Google Scholar]
- Zhang, ZL; Huang, LS; Shulmeister, VM; Chi, YI; Kim, KK; Hung, LW; Crofts, AR; Berry, EA; Kim, SH. Electron transfer by domain movement in stockbroker bc(1). Nature 1998, 392, 677–684. [Google Scholar]
- Trent, JD. A review of acquired thermotolerance, heat-shock proteins, and molecular chaperones in archaea. FEMS Microbiol. Rev 1996, 18, 249–258. [Google Scholar]
- Ellis, MJ; Knapp, S; Koeck, PJB; Fakoor-Biniaz, Z; Ladenstein, R; Hebert, H. Two-dimensional crystallization of the chaperonin TF55 from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Struct. Biol 1998, 123, 30–36. [Google Scholar]
- Grant, RA; Filman, DJ; Finkel, SE; Kolter, R; Hogle, JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol 1998, 5, 294–303. [Google Scholar]
- Zhao, G; Ceci, P; Ilari, A; Giangiacomo, L; Laue, TM; Chiancone, E; Chasteen, ND. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem 2002, 277, 27689–27696. [Google Scholar]
- Kim, S-G; Bhattacharyya, G; Grove, A; Lee, Y-H. Crystal structure of Dps-1, a functionally distinct dps protein from deinococcus radiodurans. J. Mol. Biol 2006, 361, 105–114. [Google Scholar]
- Bhattacharyya, G; Grove, A. The N-terminal extensions of deinococcus radiodurans Dps-1 mediate DNA major groove interactions as well as assembly of the dodecamer. J. Biol. Chem 2007, 282, 11921–11930. [Google Scholar]
- Ceci, P; Ilari, A; Falvo, E; Giangiacomo, L; Chiancone, E. Reassessment of protein stability, DNA binding, and protection of mycobacterium smegmatis Dps. J. Biol. Chem 2005, 280, 34776–34785. [Google Scholar]
- Andrews, SC; Smith, JMA; Hawkins, C; Williams, JM; Harrison, PM; Guest, JR. Overproduction, purification and characterization of the bacterioferritin of escherichia-coli and a C-terminally extended variant. Eur. J. Biochem 1993, 213, 329–338. [Google Scholar]
- Andrews, SC; Arosio, P; Bottke, W; Briat, JF; Vondarl, M; Harrison, PM; Laulhere, JP; Levi, S; Lobreaux, S; Yewdall, SJ. Structure, function, and evolution of ferritins. J. Inorg. Biochem 1992, 47, 161–174. [Google Scholar]
- Harrison, PM; Arosio, P. Ferritins: Molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta Bioenerg 1996, 1275, 161–203. [Google Scholar]
- Uchida, M; Klem, MT; Allen, M; Suci, P; Flenniken, M; Gillitzer, E; Varpness, Z; Liepold, LO; Young, M; Douglas, T. Biological containers: Protein cages as multifunctional nanoplatforms. Adv. Mater 2007, 19, 1025–1042. [Google Scholar]
- Destito, G; Yeh, R; Rae, CS; Finn, MG; Manchester, M. Folic acid-mediated targeting of cowpea mosaic virus particles to tumor cells. Chem. Biol 2007, 14, 1152–1162. [Google Scholar]
- Colosimo, A; Goncz, KK; Holmes, AR; Kunzelmann, K; Novelli, G; Malone, RW; Bennett, MJ; Gruenert, DC. Transfer and expression of foreign genes in mammalian cells. Biotechniques 2000, 29, 314–322. [Google Scholar]
- Zhang, N; Li, FY; Fu, QJ; Tsang, SC. Naturally occurring ferritin as a novel catalyst for selective hydroxylation of phenol. React. Kinet. Catal. Lett 2000, 71, 393–404. [Google Scholar]
- Worsdorfer, B; Woycechowsky, KJ; Hilvert, D. Directed evolution of a protein container. Science 2011, 331, 589–592. [Google Scholar]
- Andrews, SC. The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta 2010, 1800, 691–705. [Google Scholar]
- Laufberger, V. Contribution to the technique of renin seclusion. C. R. Seances. Soc. Biol. Fil 1937, 126, 107–109. [Google Scholar]
- Lawson, DM; Artymiuk, PJ; Yewdall, SJ; Smith, JMA; Livingstone, JC; Treffry, A; Luzzago, A; Levi, S; Arosio, P; Cesareni, G; et al. Solving the structure of human h-ferritin by genetically engineering intermolecular crystal contacts. Nature 1991, 349, 541–544. [Google Scholar]
- Hempstead, PD; Yewdall, SJ; Fernie, AR; Lawson, DM; Artymiuk, PJ; Rice, DW; Ford, GC; Harrison, PM. Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J. Mol. Biol 1997, 268, 424–448. [Google Scholar]
- Granier, T; Gallois, B; d’Estaintot, AD; Dautant, A; Chevelier, JM; Mellado, JM; Beaumont, C; Santambrogio, P; Arosio, P; Precigoux, G. Structure of mouse L-chain ferritin at 1.6 angstrom resolution. Acta Crystallogr. D Biol. Crystallogr 2001, 57, 1491–1497. [Google Scholar]
- Vanwuytswinkel, O; Briat, JF. Conformational changes and in vitro core-formation modifications induced by site-directed mutagenesis of the specific N-terminus of pea seed ferritin. Biochem. J 1995, 305, 959–965. [Google Scholar]
- Wardrop, AJ; Wicks, RE; Entsch, B. Occurrence and expression of members of the ferritin gene family in cowpeas. Biochem. J 1999, 337, 523–530. [Google Scholar]
- Dautant, A; Meyer, JB; Yariv, J; Precigoux, G; Sweet, RM; Kalb, AJ; Frolow, F. Structure of a monoclinic crystal form of cytochrome b1 (bacterioferritin) from E. coli. Acta Crystallogr. D Biol. Crystallogr 1998, 54, 16–24. [Google Scholar]
- Cobessi, D; Huang, LS; Ban, M; Pon, NG; Daldal, F; Berry, EA. The 2.6 angstrom resolution structure of Rhodobacter capsulatus bacterioferritin with metal-free dinuclear site and heme iron in a crystallographic “special position”. Acta Crystallogr. D Biol. Crystallogr 2002, 58, 29–38. [Google Scholar]
- Macedo, S; Romao, CV; Mitchell, E; Matias, PM; Liu, MY; Xavier, AV; LeGall, J; Teixeira, M; Lindley, P; Carrondo, MA. The nature of the di-iron site in the bacterioferritin from Desulfovibrio desulfuricans. Nat. Struct. Biol 2003, 10, 285–290. [Google Scholar]
- Theil, EC. Ferritin protein nanocages use ion channels, catalytic sites, and nucleation channels to manage iron/oxygen chemistry. Curr. Opin. Chem. Biol 2011, 15, 304–311. [Google Scholar]
- Andrews, SC. Iron storage in bacteria. Adv. Microb. Physiol 1998, 40, 281–351. [Google Scholar]
- Andrews, SC; Robinson, AK; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev 2003, 27, 215–237. [Google Scholar]
- Theil, EC. Ferritin: Structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu. Rev. Biochem 1987, 56, 289–315. [Google Scholar]
- Pettersen, EF; Goddard, TD; Huang, CC; Couch, GS; Greenblatt, DM; Meng, EC; Ferrin, TE. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem 2004, 25, 1605–1612. [Google Scholar]
- Almiron, M; Link, AJ; Furlong, D; Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 1992, 6, 2646–2654. [Google Scholar]
- Romão, CV; Mitchell, EP; McSweeney, S. The crystal structure of Deinococcus radiodurans Dps protein (DR2263) reveals the presence of a novel metal centre in the N-terminus. J. Biol. Inorg. Chem 2006, 11, 891–902. [Google Scholar]
- Bellapadrona, G; Stefanini, S; Zamparelli, C; Theil, EC; Chiancone, E. Iron translocation into and out of listeria innocua Dps and size distribution of the protein-enclosed nanomineral are modulated by the electrostatic gradient at the 3-fold “Ferritin-like” Pores. J. Biol. Chem 2009, 284, 19101–19109. [Google Scholar]
- Ilari, A; Stefanini, S; Chiancone, E; Tsernoglou, D. The dodecameric ferritin from listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct. Biol 2000, 7, 38–43. [Google Scholar]
- Chiancone, E; Ceci, P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta 2010, 1800, 798–805. [Google Scholar]
- Holm, L; Park, J. DaliLite workbench for protein structure comparison. Bioinformatics 2000, 16, 566–567. [Google Scholar]
- DaliLite Pairwise comparison of protein structures. Available online: http://www.ebi.ac.uk/Tools/dalilite/index.html (accessed on 1 June 2011).
- Jaenicke, R; Rudolph, R. Refolding and association of oligomeric proteins. Methods Enzymol 1986, 131, 218–250. [Google Scholar]
- Banyard, SH; Stammers, DK; Harrison, PM. Electron-density map of apoferritin at 2.8-a resolution. Nature 1978, 271, 282–284. [Google Scholar]
- Stefanini, S; Vecchini, P; Chiancone, E. On the mechanism of horse spleen apoferritin assembly: a sedimentation-velocity and circular-dichroism study. Biochemistry 1987, 26, 1831–1837. [Google Scholar]
- Gerl, M; Jaenicke, R. Mechanism of the self-assembly of apoferritin from horse spleen. Cross-linking and spectroscopic analysis. Eur. Biophys. J 1987, 15, 103–109. [Google Scholar]
- Gerl, M; Jaenicke, R; Smith, JMA; Harrison, PM. Self-assembly of apoferritin from horse spleen after reversible chemical modification with 2,3-Dimethylmaleic anhydride. Biochemistry 1988, 27, 4089–4096. [Google Scholar]
- Crichton, RR; Bryce, CFA. Subunit interactions in horse spleen apoferritin. Dissociation by extremes of pH. Biochem. J 1973, 133, 289–299. [Google Scholar]
- Stefanini, S; Cavallo, S; Wang, CQ; Tataseo, P; Vecchini, P; Giartosio, A; Chiancone, E. Thermal stability of horse spleen apoferritin and human recombinant H apoferritin. Arch. Biochem. Biophys 1996, 325, 58–64. [Google Scholar]
- Jin, KS; Kim, M; Rho, Y; Ahn, B; Jung, S; Kim, H; Ree, M. pH-Dependent structures of ferritin and apoferritin in solution: Disassembly and reassembly. Biomacromolecules 2011, 12, 1629–1640. [Google Scholar]
- Fan, R; Boyle, AL; Cheong, VV; Ng, SL; Orner, BP. A helix swapping study of two protein cages. Biochemistry 2009, 48, 5623–5630. [Google Scholar]
- Luzzago, A; Cesareni, G. Isolation of point mutations that affect the folding of the H-Chain of human ferritin in E. Coli. EMBO J 1989, 8, 569–576. [Google Scholar]
- Levi, S; Luzzago, A; Cesareni, G; Cozzi, A; Franceschinelli, F; Albertini, A; Arosio, P. Mechanism of ferritin iron uptake: activity of the H-Chain and deletion mapping of the ferro-oxidase site. A study of iron uptake and ferro-oxidase activity of human-liver, recombinant H-Chain ferritins, and of 2 H-Chain deletion mutants. J. Biol. Chem 1988, 263, 18086–18092. [Google Scholar]
- Levi, S; Luzzago, A; Franceschinelli, F; Santambrogio, P; Cesareni, G; Arosio, P. Mutational analysis of the channel and loop sequences of human ferritin H-Chain. Biochem. J 1989, 264, 381–388. [Google Scholar]
- Ingrassia, R; Gerardi, G; Biasiotto, G; Arosio, P. Mutations of ferritin H chain C-terminus produced by nucleotide insertions have altered stability and functional properties. J. Biochem 2006, 139, 881–885. [Google Scholar]
- Yoshizawa, K; Mishima, Y; Park, SY; Heddle, JG; Tame, JRH; Iwahori, K; Kobayashi, M; Yamashita, I. Effect of N-terminal residues on the structural stability of recombinant horse L-chain apoferritin in an acidic environment. J. Biochem 2007, 142, 707–713. [Google Scholar]
- Santambrogio, P; Pinto, P; Levi, S; Cozzi, A; Rovida, E; Albertini, A; Artymiuk, P; Harrison, PM; Arosio, P. Effects of modifications near the 2-, 3- and 4-fold symmetry axes an human ferritin renaturation. Biochem. J 1997, 322, 461–468. [Google Scholar]
- Kilic, MA; Spiro, S; Moore, GR. Stability of a 24-meric homopolymer: Comparative studies of assembly-defective mutants of Rhodobacter capsulatus bacterioferritin and the native protein. Protein Sci 2003, 12, 1663–1674. [Google Scholar]
- Frolow, F; Kalb, AJ; Yariv, J. Structure of a unique twofold symmetrical heme-binding site. Nat. Struct. Biol 1994, 1, 453–460. [Google Scholar]
- Zhang, Y; Raudah, S; Teo, H; Teo, GWS; Fan, RL; Sun, XM; Orner, BP. Alanine-shaving mutagenesis to determine key interfacial residues governing the assembly of a Nano-cage Maxi-ferritin. J. Biol. Chem 2010, 285, 12078–12086. [Google Scholar]
- Roy, S; Gupta, S; Das, S; Sekar, K; Chatterji, D; Vijayan, M. X-ray analysis of mycobacterium smegmatis dps and a comparative study involving other Dps and Dps-like molecules. J. Mol. Biol 2004, 339, 1103–1113. [Google Scholar]
- Gupta, S; Chatterji, D. Bimodal protection of DNA by mycobacterium smegmatis DNA-binding protein from stationary phase cells. J. Biol. Chem 2003, 278, 5235–5241. [Google Scholar]
- Kiessling, LL; Gestwicki, JE; Strong, LE. Synthetic multivalent ligands as probes of signal transduction. Angew. Chem. Int. Ed. Engl 2006, 45, 2348–2368. [Google Scholar]
- Roy, S; Saraswathi, R; Gupta, S; Sekar, K; Chatterji, D; Vijayan, M. Role of N and C-terminal tails in DNA binding and assembly in Dps: Structural studies of mycobacterium smegmatis Dps deletion mutants. J. Mol. Biol 2007, 370, 752–767. [Google Scholar]
- Chowdhury, RP; Saraswathi, R; Chatterji, D. Mycobacterial stress regulation: The dps “Twin Sister” defense mechanism and structure-function relationship. IUBMB Life 2010, 62, 67–77. [Google Scholar]
- Chowdhury, RP; Vijayabaskar, MS; Vishveshwara, S; Chatterji, D. Molecular mechanism of in vitro oligomerization of Dps from Mycobacterium smegmatis: Mutations of the residues identified by “interface cluster” analysis. Biochemistry 2008, 47, 11110–11117. [Google Scholar]
- Chiaraluce, R; Consalvi, V; Cavallo, S; Ilari, A; Stefanini, S; Chiancone, E. The unusual dodecameric ferritin from listeria innocua dissociates below pH 2.0. Eur. J. Biochem 2000, 267, 5733–5741. [Google Scholar]





© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhang, Y.; Orner, B.P. Self-Assembly in the Ferritin Nano-Cage Protein Superfamily. Int. J. Mol. Sci. 2011, 12, 5406-5421. https://doi.org/10.3390/ijms12085406
Zhang Y, Orner BP. Self-Assembly in the Ferritin Nano-Cage Protein Superfamily. International Journal of Molecular Sciences. 2011; 12(8):5406-5421. https://doi.org/10.3390/ijms12085406
Chicago/Turabian StyleZhang, Yu, and Brendan P. Orner. 2011. "Self-Assembly in the Ferritin Nano-Cage Protein Superfamily" International Journal of Molecular Sciences 12, no. 8: 5406-5421. https://doi.org/10.3390/ijms12085406