KCNQ1 Haplotypes Associate with Type 2 Diabetes in Malaysian Chinese Subjects
Abstract
:1. Introduction
2. Results
2.1. Association of KCNQ1 SNPs with T2D
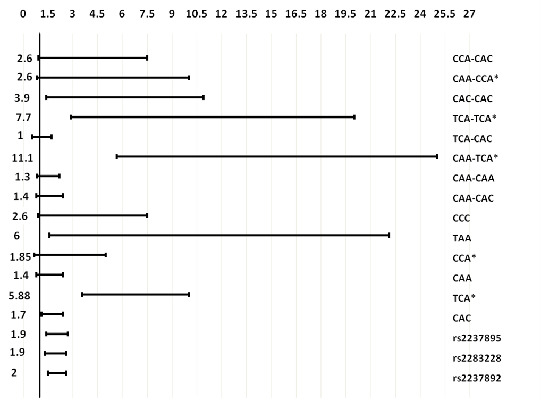
2.2. Association of KCNQ1 Haplotypes and Diplotypes with T2D
2.3. Impact of KCNQ1 SNPs, Haplotypes and Diplotypes on Beta-Cell Function in Normal Subjects
3. Discussion
4. Materials and Methods
4.1. Subjects and Data Collection
4.2. Biochemical Analyses
4.3. Genetic Analyses
4.4. Statistical Analysis
5. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Neyroud, N; Richard, P; Vignier, N; Donger, C; Denjoy, I; Demay, L; Shkolnikova, M; Pesce, R; Chevalier, P; Hainque, B; et al. Genomic organization of the KCNQ1 K+ channel gene and identification of C-terminal mutations in the long-QT syndrome. Circ Res 1999, 84, 290–297. [Google Scholar]
- Barhanin, J; Lesage, F; Guillemare, E; Fink, M; Lazdunski, M; Romey, G. KVLQT1 and lsK (minK) proteins associate to form the IKScardiac potassium current. Nature 1996, 384, 78–80. [Google Scholar]
- Thevenod, F. Ion channels in secretory granules of the pancreas and their role in exocytosis and release of secretory proteins. Am J Physiol Cell Physiol 2002, 283, C651–C672. [Google Scholar]
- Ullrich, S; Su, J; Ranta, F; Wittekindt, OH; Ris, F; Rosler, M; Gerlach, U; Heitzmann, D; Warth, R; Lang, F. Effects of IKs channel inhibitors in insulin-secreting INS-1 cells. Pflüg Arch Eur J Physiol 2005, 451, 428–436. [Google Scholar]
- Prokopenko, I; McCarthy, MI; Lindgren, CM. Type 2 diabetes: New genes, new understanding. Trends Genet 2008, 24, 613–621. [Google Scholar]
- Unoki, H; Takahashi, A; Kawaguchi, T; Hara, K; Horikoshi, M; Andersen, G; Ng, DP; Holmkvist, J; Borch-Johnsen, K; Jorgensen, T; et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008, 40, 1098–1102. [Google Scholar]
- Yasuda, K; Miyake, K; Horikawa, Y; Hara, K; Osawa, H; Furuta, H; Hirota, Y; Mori, H; Jonsson, A; Sato, Y; et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008, 40, 1092–1097. [Google Scholar]
- Tsai, FJ; Yang, CF; Chen, CC; Chuang, LM; Lu, CH; Chang, CT; Wang, TY; Chen, RH; Shiu, CF; Liu, YM; et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet 2010, 6. [Google Scholar] [CrossRef]
- Voight, BF; Scott, LJ; Steinthorsdottir, V; Morris, AP; Dina, C; Welch, RP; Zeggini, E; Huth, C; Aulchenko, YS; Thorleifsson, G; et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010, 42, 579–589. [Google Scholar]
- Hu, C; Wang, C; Zhang, R; Ma, X; Wang, J; Lu, J; Qin, W; Bao, Y; Xiang, K; Jia, W. Variations in KCNQ1 are associated with type 2 diabetes and beta cell function in a Chinese population. Diabetologia 2009, 52, 1322–1325. [Google Scholar]
- Liu, Y; Zhou, DZ; Zhang, D; Chen, Z; Zhao, T; Zhang, Z; Ning, M; Hu, X; Yang, YF; Zhang, ZF; et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia 2009, 52, 1315–1321. [Google Scholar]
- Qi, Q; Li, H; Loos, RJ; Liu, C; Wu, Y; Hu, FB; Wu, H; Lu, L; Yu, Z; Lin, X. Common variants in KCNQ1 are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Hum Mol Genet 2009, 18, 3508–3515. [Google Scholar]
- Tan, JT; Nurbaya, S; Gardner, D; Ye, S; Tai, ES; Ng, DP. Genetic variation in KCNQ1 associates with fasting glucose and beta-cell function: a study of 3,734 subjects comprising three ethnicities living in Singapore. Diabetes 2009, 58, 1445–1449. [Google Scholar]
- Tan, JT; Ng, DP; Nurbaya, S; Ye, S; Lim, XL; Leong, H; Seet, LT; Siew, WF; Kon, W; Wong, TY; et al. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab 2010, 95, 390–397. [Google Scholar]
- Been, LF; Ralhan, S; Wander, GS; Mehra, NK; Singh, JR; Mulvihill, JJ; Aston, CE; Sanghera, DK. Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: A study of 3,310 subjects from India and the US. BMC Med Genet 2011, 12. [Google Scholar] [CrossRef]
- Rees, SD; Hydrie, MZ; Shera, AS; Kumar, S; O’Hare, JP; Barnett, AH; Basit, A; Kelly, MA. Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia 2011, 54, 1368–1374. [Google Scholar]
- Holmkvist, J; Banasik, K; Andersen, G; Unoki, H; Jensen, TS; Pisinger, C; Borch-Johnsen, K; Sandbaek, A; Lauritzen, T; Brunak, S; et al. The type 2 diabetes associated minor allele of rs2237895 KCNQ1 associates with reduced insulin release following an oral glucose load. PLoS One 2009, 4. [Google Scholar] [CrossRef]
- Jonsson, A; Isomaa, B; Tuomi, T; Taneera, J; Salehi, A; Nilsson, P; Groop, L; Lyssenko, V. A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes 2009, 58, 2409–2413. [Google Scholar]
- Alberti, KG; Eckel, RH; Grundy, SM; Zimmet, PZ; Cleeman, JI; Donato, KA; Fruchart, JC; James, WP; Loria, CM; Smith, SC, Jr. Harmonizing the metabolic syndrome: A joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009, 120, 1640–1645. [Google Scholar]
- Xu, M; Bi, Y; Xu, Y; Yu, B; Huang, Y; Gu, L; Wu, Y; Zhu, X; Li, M; Wang, T; et al. Combined effects of 19 common variations on type 2 diabetes in Chinese: results from two community-based studies. PLoS One 2010, 5. [Google Scholar] [CrossRef]
- Lee, YH; Kang, ES; Kim, SH; Han, SJ; Kim, CH; Kim, HJ; Ahn, CW; Cha, BS; Nam, M; Nam, CM; et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Hum Genet 2008, 53, 991–998. [Google Scholar]
- Warth, R; Garcia Alzamora, M; Kim, JK; Zdebik, A; Nitschke, R; Bleich, M; Gerlach, U; Barhanin, J; Kim, SJ. The role of KCNQ1/KCNE1 K+ channels in intestine and pancreas: lessons from the KCNE1 knockout mouse. Pflüg Arch Eur J Physiol 2002, 443, 822–828. [Google Scholar]
- Boini, KM; Graf, D; Hennige, AM; Koka, S; Kempe, DS; Wang, K; Ackermann, TF; Foller, M; Vallon, V; Pfeifer, K; et al. Enhanced insulin sensitivity of gene-targeted mice lacking functional KCNQ1. Am J Physiol Regul Integr Comp Physiol 2009, 296, R1695–R1701. [Google Scholar]
- The United States National Library of Medicine. Available online: http://www.ncbi.nlm.nih.gov/snp (assessed on 22 February 2010).
- Institute of Human Genetics, Technical University Munich; Munich, Germany. Available online: http://ihg.gsf.de/cgi-bin/hw/hwa1.pl (accessed on 6 August 2011).


| Parameters | Normal n = 230 | Type 2 diabetes n = 300 | P-value | |
|---|---|---|---|---|
| Gender | Male % | 61.3 | 51 | |
| Female % | 38.7 | 49 | ||
| Family history of diabetes % | Male % | 32 | 76 | |
| Female % | 34 | 63 | ||
| Age (years) | 52.9 ± 9.15 | 49.8 ± 7.42 | <0.001 | |
| Height (m) | 1.62 ± 0.08 | 1.63 ± 0.09 | 0.18 | |
| Weight (kg) | 63.1 ± 15.5 | 75.0 ± 15.2 | <0.001 | |
| BMI(kg/m2) | 24.1 ± 5.14 | 28.3 ± 5.15 | <0.001 | |
| Waist (cm) | 84.7 ± 13.1 | 95.7 ± 12.3 | <0.001 | |
| Hip (cm) | 99 ± 10.1 | 104 ± 10.1 | 0.001 | |
| Waist/Hip | 0.85 ± 0.07 | 0.92 ± 0.07 | <0.001 | |
| Systolic blood pressure | 136 ± 18.5 | 136 ± 18.5 | 0.97 | |
| 81 ± 9.7 | 82 ± 10.5 | 0.20 | ||
| Fasting insulin (pmol/L) | 63.8 ± 44.6 | 103 ± 56.6 | <0.001 | |
| Fasting glucose (mmol/L) | 5.1 ± 0.49 | 8.3 ± 2.74 | <0.001 | |
| Triacylglycerol (mmol/L) | 1.1 ± 0.44 | 2.1 ± 1.20 | <0.001 | |
| HDL cholesterol (mmol/L) | 1.52 ± 0.32 | 1.21 ± 0.28 | <0.001 | |
| Insulin resistance (IR) | 1.4 ± 0.93 | 2.5 ± 1.44 | <0.001 | |
| Recessive model | Dominant model | Additive model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NPs | Group | Genotype n (distribution %) | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | ||
| First logistic regression model, analysis adjusted for age and gender
| ||||||||||
| rs2237892 | TT | CT | CC | 1.9 (1.29–2.66) | 0.001 | 2.1 (1.09–3.94) | 0.026 | 1.6 (1.25–2.18) | 0.0005 | |
| normal | 27(11.7) | 90(39.1) | 113(49.1) | |||||||
| diabetic | 18(6.0) | 99(33.0) | 183(61.0) | |||||||
| rs2283228 | CC | AC | AA | 1.8 (1.24–2.52) | 0.002 | 1.7 (0.97–2.93) | 0.06 | 1.5 (1.17–1.97) | 0.002 | |
| normal | 33(14.3) | 98(42.6) | 99(43.0) | |||||||
| diabetic | 27(9.0) | 105(35.0) | 168(56.0) | |||||||
| rs2237895 | CC | AC | AA | 1.6 (0.83–3.17) | 0.16 | 1.7 (1.17–2.39) | 0.004 | 1.5 (1.14–2.01) | 0.004 | |
| normal | 14(6.1) | 96(41.7) | 120(52.2) | |||||||
| diabetic | 30(10.0) | 147(49.0) | 123(41.0) | |||||||
| Second logistic regression model, analysis adjusted for age, gender, and body mass index
| ||||||||||
| rs2237892 * | TT | CT | CC | 2.2 (1.5–3.4) | 0.0002 | 2.7 (1.3–5.5) | 0.007 | 2.0 (1.4–2.7) | 5.1 × 10−5 | |
| normal | 27(12.6) | 81(37.7) | 107(49.8) | |||||||
| diabetic | 18(6.0) | 99(33.0 | 183(61.0) | |||||||
| rs2283228 * | CC | AC | AA | 2.3 (1.5–3.5) | 7.3 × 10−5 | 2.2 (1.2–4.1) | 0.011 | 1.9 (1.4–2.5) | 5.2 × 10−5 | |
| normal | 33(14.9) | 89(40.3) | 99(44.8) | |||||||
| diabetic | 27(9.0) | 105(35.0) | 168(56.0) | |||||||
| rs2237895 * | CC | AC | AA | 3.7 (1.7–8.1) | 0.001 | 2.0 (1.3–3.0) | 0.001 | 1.9 (1.4–2.7) | 7.8 × 10−5 | |
| normal | 11(5.0) | 96(44.0) | 111(50.9) | |||||||
| diabetic | 30(10.0) | 147(49.0) | 123(41.0) | |||||||
| Haplotypes rs2237892, rs2283228, rs2237895 | Frequency
| |||||
|---|---|---|---|---|---|---|
| Normal n (230) | Type 2 diabetes n (300) | Odds ratio | 95% CI | P-Value | Overall P-value | |
| First logistic regression model, analysis adjusted for age and gender
| ||||||
| CAC | 0.44 | 0.51 | 1.4 | 0.99–2.01 | 0.057 | 7.49 × 10−6 a |
| TCA | 0.33 | 0.15 | 0.33 | 0.21–0.51 | 8.4 × 10−7 | |
| CAA | 0.13 | 0.16 | 1.3 | 0.86–2.21 | 0.263 | |
| CCA | 0.05 | 0.04 | 0.7 | 0.30–1.63 | 0.41 | |
| TAA | 0.01 | 0.06 | 4.6 | 1.31–16.19 | 0.017 | |
| CCC | 0.02 | 0.07 | 3.2 | 1.20–8.81 | 0.021 | |
| Second logistic regression model, analysis adjusted for age, gender and body mass index
| ||||||
| CAC * | 0.44 | 0.51 | 1.7 | 1.1–2.4 | 0.008 | |
| TCA * | 0.34 | 0.15 | 0.17 | 0.1–0.28 | 3.7 × 10−11 | |
| CAA * | 0.14 | 0.16 | 1.4 | 0.79–2.4 | 0.26 | |
| CCA * | 0.05 | 0.04 | 0.54 | 0.2–1.5 | 0.23 | |
| TAA * | 0.01 | 0.06 | 6.0 | 1.6–22.1 | 0.007 | |
| CCC * | 0.02 | 0.07 | 2.6 | 0.88–7.5 | 0.083 | |
| Diplotypes rs2237892, rs2283228, rs2237895 | (Frequency)
| |||||
|---|---|---|---|---|---|---|
| Normal n (230) | Type 2 diabetes n (300) | Odds ratio | 95% CI | P-Value | Overall P-value | |
| First logistic regression model, analysis adjusted for age and gender
| ||||||
| CAA-CAC | 0.22 | 0.25 | 1.3 | 0.86–1.99 | 0.21 | 8.1 × 10−7 a |
| CAA-CAA | 0.13 | 0.16 | 1.3 | 0.81–2.21 | 0.26 | |
| CAA-TCA | 0.18 | 0.09 | 0.4 | 0.21–0.63 | 0.0003 | |
| TCA-CAC | 0.13 | 0.12 | 0.92 | 0.54–1.57 | 0.77 | |
| TCA-TCA | 0.12 | 0.02 | 0.16 | 0.06–0.40 | 8 × 10−5 | |
| CAC-CAC | 0.03 | 0.06 | 2.2 | 0.85–5.74 | 0.104 | |
| CAA-CCA | 0.05 | 0.03 | 0.48 | 0.19–1.19 | 0.11 | |
| CCA-CAC | 0.04 | 0.06 | 1.8 | 0.78–4.10 | 0.17 | |
| Second logistic regression model, analysis adjusted for age, gender, and body mass index
| ||||||
| CAA-CAC * | 0.23 | 0.25 | 1.4 | 0.79–2.4 | 0.26 | |
| CAA-CAA * | 0.14 | 0.16 | 1.3 | 0.82–1.3 | 0.26 | |
| CAA-TCA * | 0.19 | 0.08 | 0.09 | 0.04–0.18 | 2.9 × 10−10 | |
| TCA-CAC * | 0.14 | 0.12 | 1.0 | 0.54–1.7 | 0.88 | |
| TCA-TCA * | 0.12 | 0.02 | 0.13 | 0.05–0.34 | 4.4 × 10−5 | |
| CAC-CAC * | 0.01 | 0.06 | 3.9 | 1.4–10.9 | 0.008 | |
| CAA-CCA * | 0.05 | 0.03 | 0.38 | 0.12–1.2 | 0.09 | |
| CCA-CAC * | 0.03 | 0.06 | 2.6 | 0.92–7.5 | 0.07 | |
| Non adjusted HOMA-B means(CI) | Adjusted for age, gender | Adjusted for age, gender, BMI | |||||
|---|---|---|---|---|---|---|---|
| HOMA-B mean(CI) | Parameter estimate P-value | Univariate P-value | HOMA-B Mean(CI) | Parameter estimate P-value | Univariate P-value | ||
| rs2237892 | |||||||
| CC(n = 113) | 96(90–103) | 98(92–105) | 0.006 | 0.002 | 100(95–106) | 0.01 | 0.0002 |
| CT(n = 90) | 94(87–101) | 92(85–98) | 0.0004 | 90(85–96) | 0.0003 | ||
| TT(n = 27) Ref | 120(104–137) | 121(106–138) | 118(105–132) | ||||
| rs2283228 | |||||||
| AA(99) | 97(90–104) | 97(90–104) | 0.33 | 0.584 | 102(96–109) | 0.80 | 0.034 |
| AC(98) | 97(87–101) | 97(90–103) | 0.30 | 92(86–97) | 0.039 | ||
| CC(33) Ref | 104(92–118) | 104(92–118) | 104(94–115) | ||||
| rs2237895 | |||||||
| CC(14) | 100(83–122) | 98(81–119) | 0.91 | 0.74 | 106(90–125) | 0.36 | 0.554 |
| AC(96) | 95(88–102) | 96(89–103) | 0.44 | 96(91–103) | 0.70 | ||
| AA(120) Ref | 100(94–107) | 99(93–106) | 98(93–104) | ||||
| haplotypes | |||||||
| CAA(n = 30) | 84(74–95) | 85(75–97) | 0.001 | 0.003 | 90(81–100) | 0.007 | 0.014 |
| CAC(n = 102) | 97(91–104) | 97(91–104) | 0.026 | 97(92–103) | 0.026 | ||
| TCA(n = 75) Ref | 111(103–120) | 110(102–119) | 107(101–115) | ||||
| Diplotype | |||||||
| CAA-CAA(n = 30) | 84(75–94) | 85(76–95) | 4.0 × 10−5 | 1.5 × 10−6 | 90(81–99) | 0.0001 | 5.7 × 10−6 |
| CAA-CAC(n = 51) | 106(97–115) | 107(98–116) | 0.12 | 110(102–119) | 0.20 | ||
| CAA-TCA(n = 42) | 117(107–129) | 114(104–126) | 0.55 | 105(96–115) | 0.062 | ||
| TCA-CAC(n = 30) | 83(75–93) | 84(75–94) | 1.9 × 10−5 | 85(77–94) | 5.8 × 10−6 | ||
| TCA-TCA(n = 27) Ref | 120(106–134) | 120(107–135) | 120(108–133) | ||||
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Saif-Ali, R.; Ismail, I.S.; Al-Hamodi, Z.; Al-Mekhlafi, H.M.; Siang, L.C.; Alabsi, A.M.; Muniandy, S. KCNQ1 Haplotypes Associate with Type 2 Diabetes in Malaysian Chinese Subjects. Int. J. Mol. Sci. 2011, 12, 5705-5718. https://doi.org/10.3390/ijms12095705
Saif-Ali R, Ismail IS, Al-Hamodi Z, Al-Mekhlafi HM, Siang LC, Alabsi AM, Muniandy S. KCNQ1 Haplotypes Associate with Type 2 Diabetes in Malaysian Chinese Subjects. International Journal of Molecular Sciences. 2011; 12(9):5705-5718. https://doi.org/10.3390/ijms12095705
Chicago/Turabian StyleSaif-Ali, Riyadh, Ikram S. Ismail, Zaid Al-Hamodi, Hesham M. Al-Mekhlafi, Lee C. Siang, Aied M. Alabsi, and Sekaran Muniandy. 2011. "KCNQ1 Haplotypes Associate with Type 2 Diabetes in Malaysian Chinese Subjects" International Journal of Molecular Sciences 12, no. 9: 5705-5718. https://doi.org/10.3390/ijms12095705