Phorbol Esters Isolated from Jatropha Meal Induced Apoptosis-Mediated Inhibition in Proliferation of Chang and Vero Cell Lines
Abstract
:Introduction
2. Results and Discussion
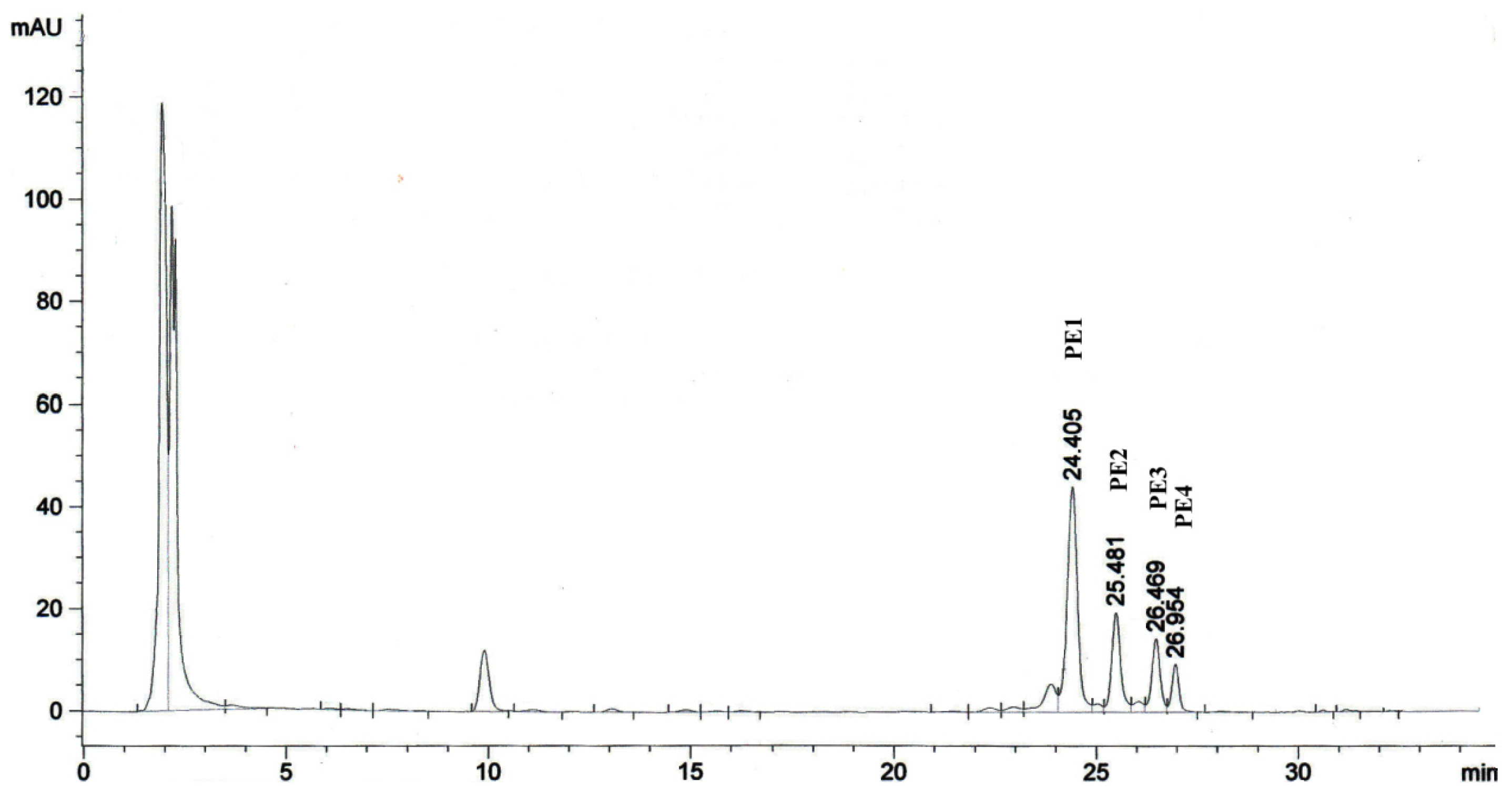
2.1. Isolation of Phorbol Esters
2.2. Proliferation Assay
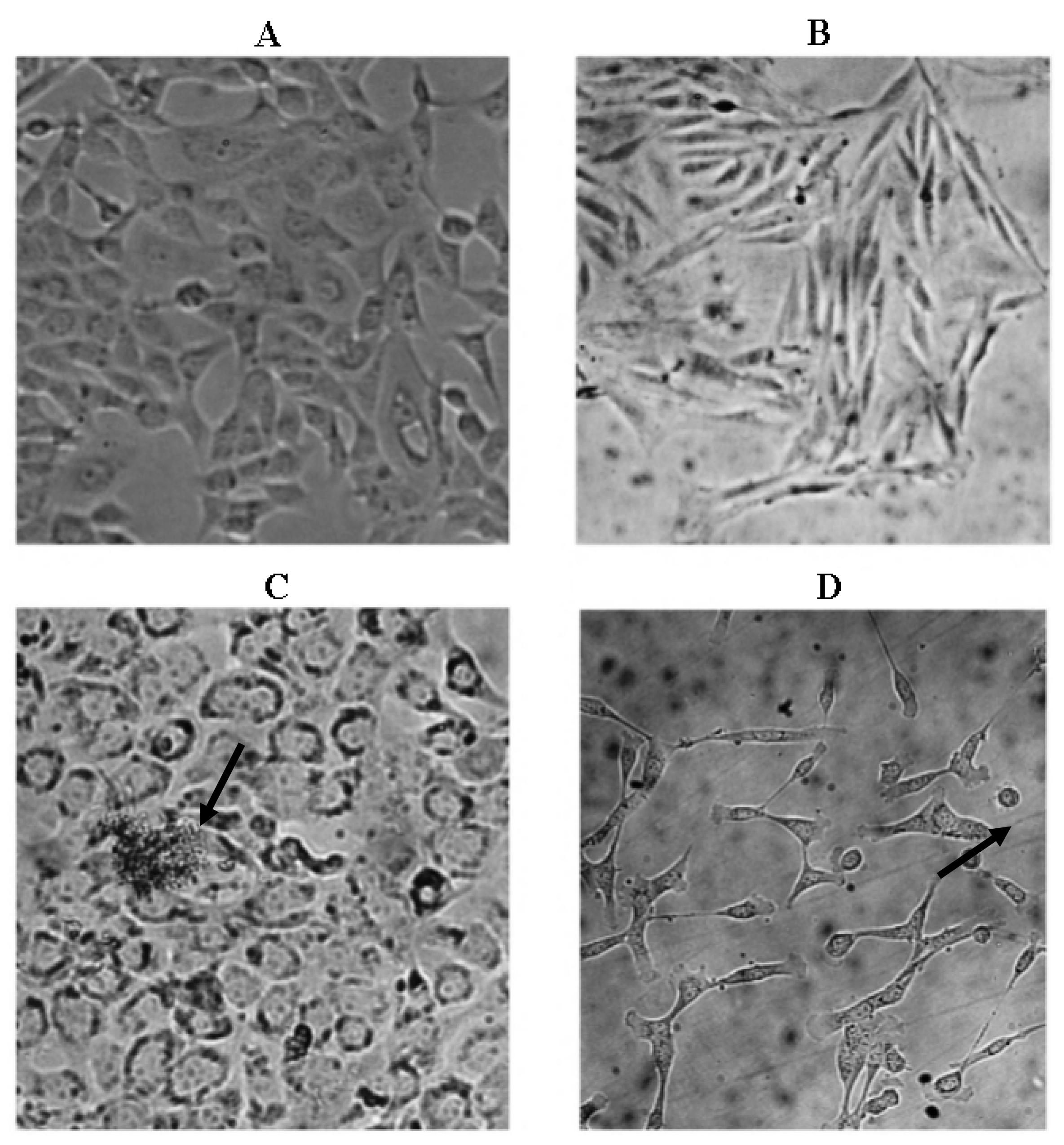
2.3. Microscopic Examination
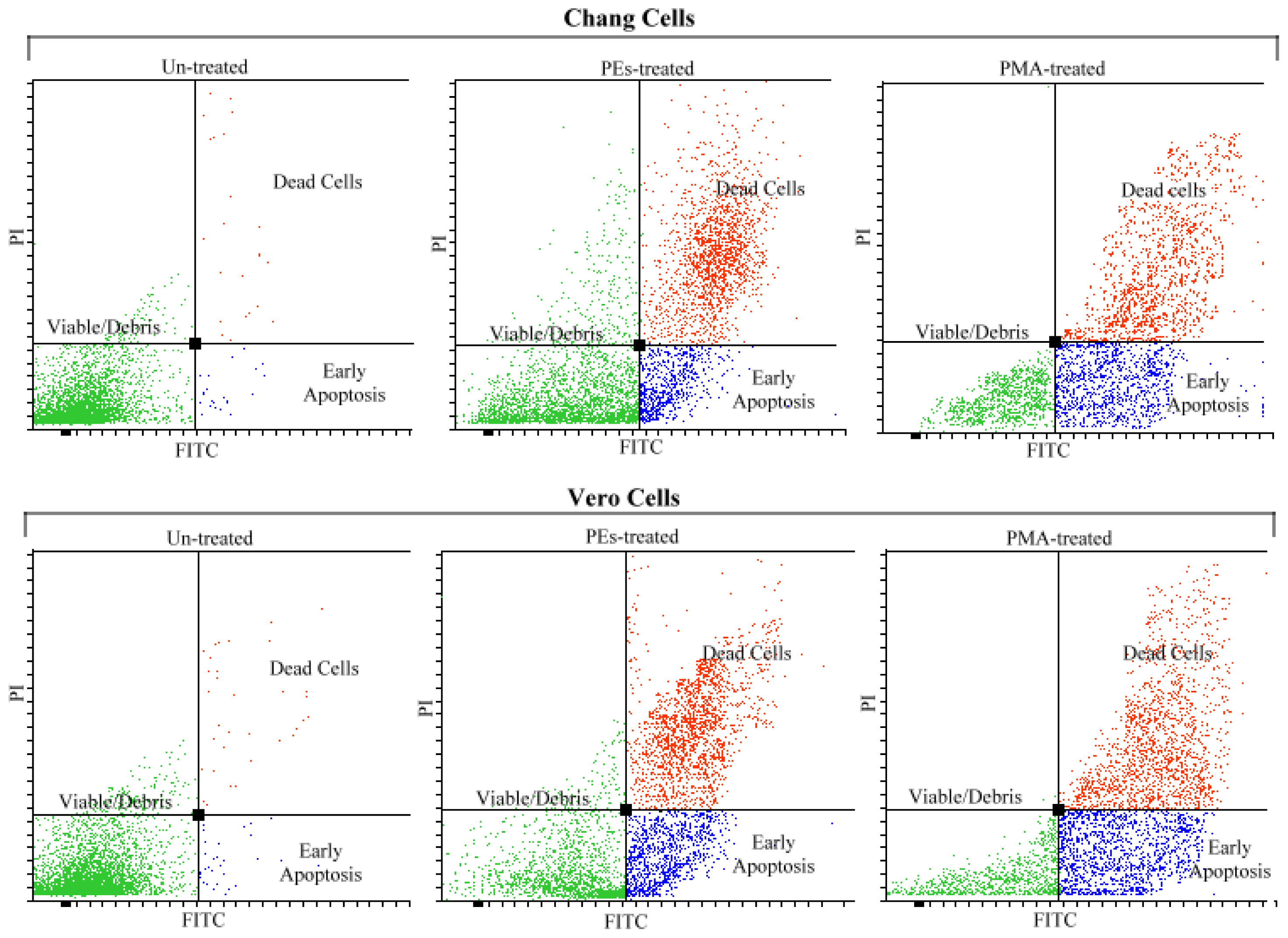
2.4. Analysis of Apoptosis by Flow Cytometry
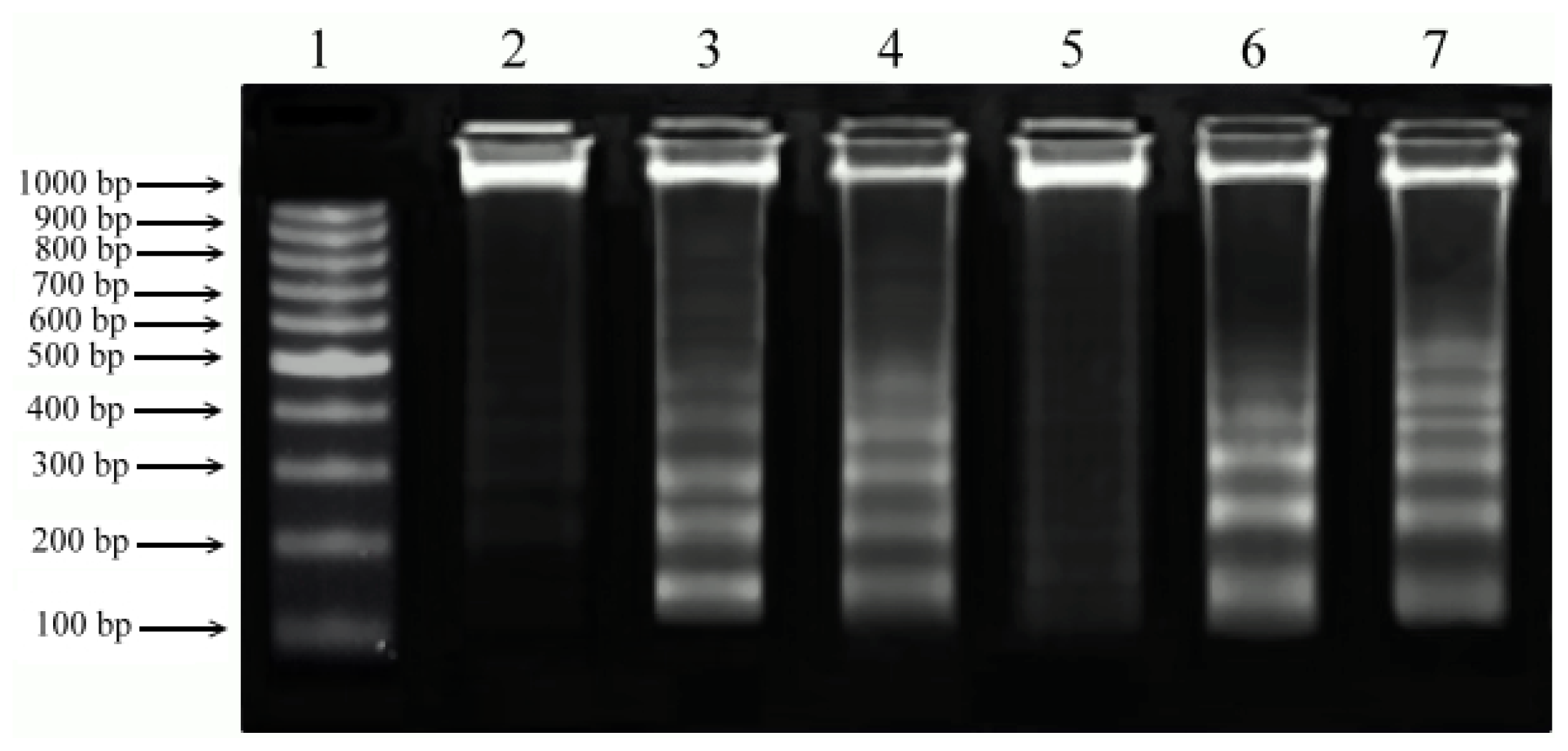
2.5. DNA Fragmentation
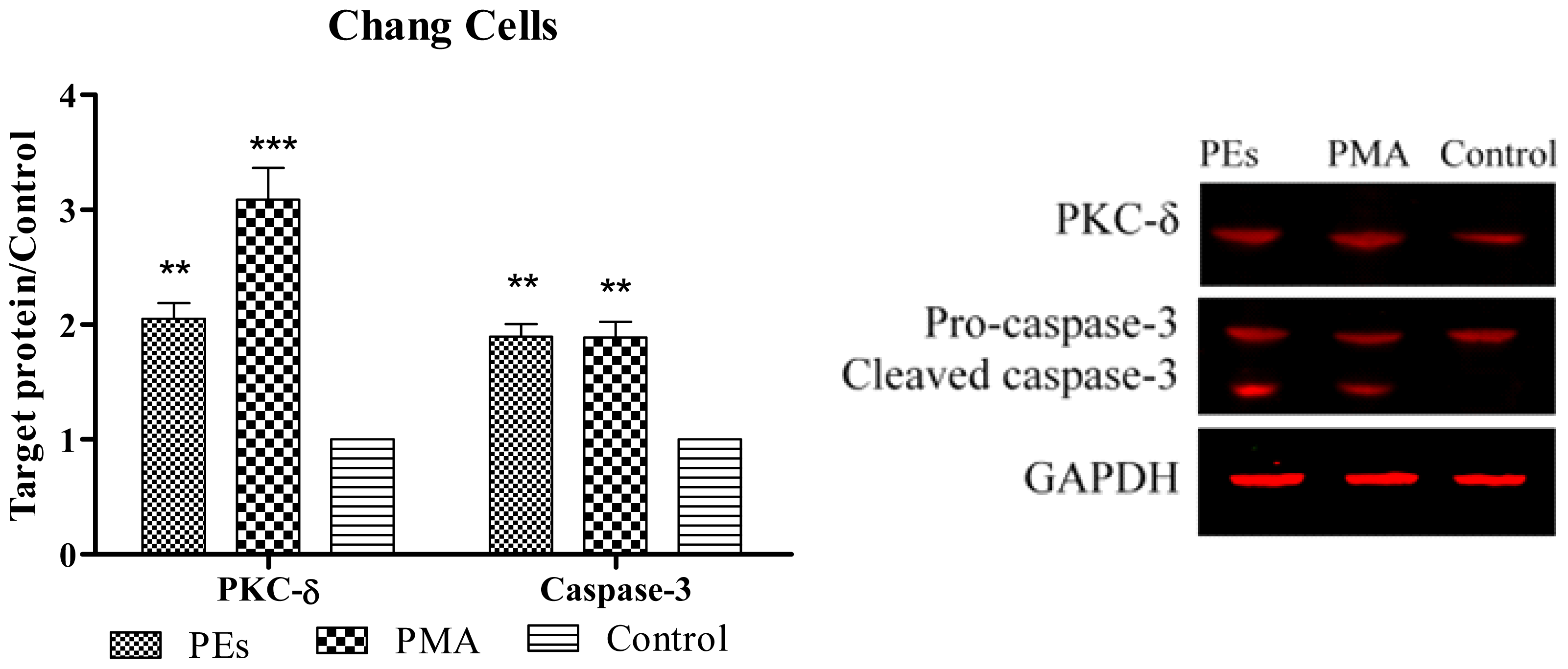
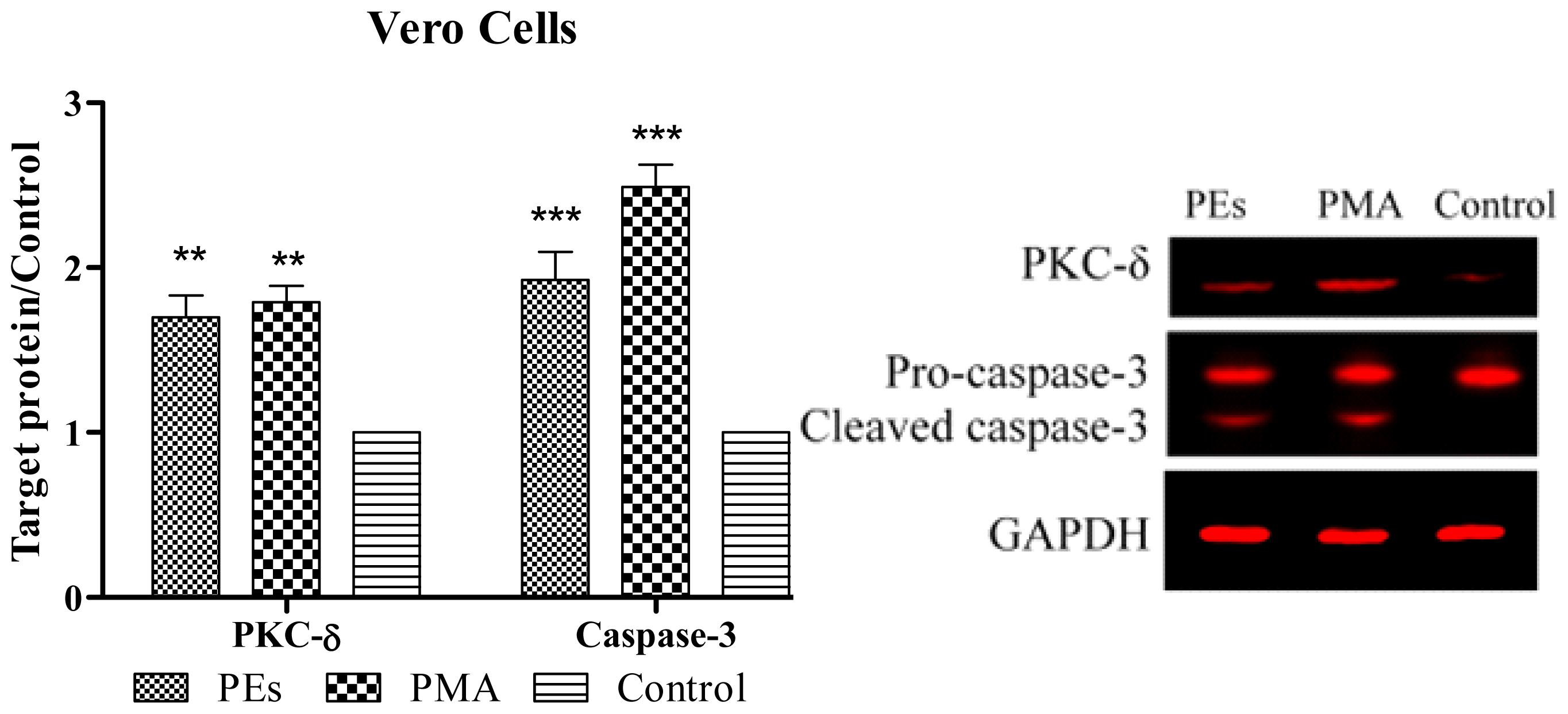
2.6. Western Blot Assay
3. Experimental Section
3.1. Plant Materials
3.2. Phorbol Esters Isolation
3.3. Cell Lines and Cell Culture
3.4. Proliferation Assay
3.5. Microscopic Examination
3.6. Analysis of Apoptosis by Flow-Cytometry
3.7. DNA Fragmentation Assay
3.8. Western Blotting
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
References
- Murugesan, A.; Umarani, C.; Subramanian, R.; Nedunchezhian, N. Bio-diesel as an alternative fuel for diesel engines—A review. Renew. Sust. Energ. Rev 2009, 13, 653–662. [Google Scholar]
- Oskoueian, E.; Abdullah, N.; Saad, W.Z.; Omar, A.R.; Puteh, M.B.; Ho, Y.W. Anti-nutritional metabolites and effect of treated Jatropha curcas kernel meal on rumen fermentation in vitro. J. Anim. Vet. Adv 2011, 10, 214–220. [Google Scholar]
- King, A.J.; He, W.; Cuevas, J.A.; Freudenberger, M.; Ramiaramanana, D.; Graham, I.A. Potential of Jatropha curcas as a source of renewable oil and animal feed. J. Exp. Bot 2009, 60, 2897–2905. [Google Scholar]
- Devappa, R.K. Isolation, Characterization and Potential Agro-Pharmaceutical Applications of Phorbol Esters from Jatropha curcas Oil. Ph.D. Thesis, University of Hohenheim, Stuttgart, Germany, Ferburay 2012. [Google Scholar]
- Rakshit, K.D.; Darukeshwara, J.; Rathina Raj, K.; Narasimhamurthy, K.; Saibaba, P.; Bhagya, S. Toxicity studies of detoxified Jatropha meal (Jatropha curcas) in rats. Food Chem. Toxicol 2008, 46, 3621–3625. [Google Scholar]
- Abdel-Hafez, A.; Nakamura, N.; Hattori, M. Biotransformation of phorbol by human intestinal bacteria. Chem. Pharm. Bull 2002, 50, 160–164. [Google Scholar]
- Griner, E.M.; Kazanietz, M.G. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer 2007, 7, 281–294. [Google Scholar]
- Hatton, J.P.; Gaubert, F.; Cazenave, J.P.; Schmitt, D. Microgravity modifies protein kinase C isoform translocation in the human monocytic cell line U937 and human peripheral blood T-cells. J. Cell Biochem 2002, 87, 39–50. [Google Scholar]
- Goel, G.; Makkar, H.; Francis, G.; Becker, K. Phorbol esters: Structure, biological activity, and toxicity in animals. Int. J. Toxicol 2007, 26, 279–288. [Google Scholar]
- Makkar, H.P.S.; Siddhuraju, S.; Siddhuraju, P.; Becker, K. Plant Secondary Metabolites; Humana Press: New York, NY, USA, 2007; pp. 101–106. [Google Scholar]
- Li, C.Y.; Devappa, R.K.; Liu, J.X.; Lv, J.M.; Makkar, H.P.S.; Becker, K. Toxicity of Jatropha curcas phorbol esters in mice. Food Chem. Toxicol 2010, 48, 620–625. [Google Scholar]
- Haas, W.; Sterk, H.; Mittelbach, M. Novel 12-deoxy-16-hydroxyphorbol diesters isolated from the seed oil of Jatropha curcas. J. Nat. Prod 2002, 65, 1434–1440. [Google Scholar]
- Roach, J.S.; Devappa, R.K.; Makkar, H.P.S.; Becker, K. Isolation, stability and bioactivity of Jatropha curcas phorbol esters. Fitoterapia 2012, 83, 586–592. [Google Scholar]
- Mentlein, R. The tumor promoter 12-O-tetradecanoyl phorbol 13-acetate and regulatory diacylglycerols are substrates for the same carboxylesterase. J. Biol. Chem 1986, 261, 7816–7818. [Google Scholar]
- Bond, J.A.; Gescher, A.J.; Verschoyle, R.D.; Lemoine, N.R.; Errington, R.; Wiltshire, M.; Smith, P.J.; Thomas, D.W. Cytotoxic action of phorbol esters on human pancreatic cancer cells. Int. J. Cancer 2007, 121, 1445–1454. [Google Scholar]
- Halaweish, F.T.; Kronberg, S.; Hubert, M.B.; Rice, J.A. Toxic and aversive diterpenes of Euphorbia esula. J. Chem. Ecol 2002, 28, 1599–1611. [Google Scholar]
- Kazanietz, M.G. Targeting protein kinase C and “non-kinase” phorbol ester receptors: Emerging concepts and therapeutic implications. Biochim. Biophys. Acta Protein Proteonomics 2005, 1754, 296–304. [Google Scholar]
- Gobbi, G.; Mirandola, P.; Carubbi, C.; Micheloni, C.; Malinverno, C.; Lunghi, P.; Bonati, A.; Vitale, M. Phorbol ester-induced PKC down-modulation sensitizes AML cells to TRAIL-induced apoptosis and cell differentiation. Blood 2009, 113, 3080–3087. [Google Scholar]
- Von Burstin, V.A.; Xiao, L.; Kazanietz, M.G. Bryostatin 1 Inhibits phorbol ester-induced apoptosis in prostate cancer cells by differentially modulating protein kinase C (PKC) Δ translocation and preventing PKC Δ-Mediated release of tumor necrosis factor-α. Mol. Pharmacol 2010, 78, 325–332. [Google Scholar]
- Shrotriya, S.; Kundu, J.K.; Na, H.K.; Surh, Y.J. Diallyl trisulfide inhibits phorbol ester-induced tumor promotion, activation of AP-1, and expression of COX-2 in mouse skin by blocking JNK and AKT signaling. Cancer Res 2010, 70, 1932–1940. [Google Scholar]
- Ikuta, S.; Edamatsu, H.; Li, M.; Hu, L.; Kataoka, T. Crucial role of phospholipase Cɛ in skin inflammation induced by tumor-promoting phorbol ester. Cancer Res 2008, 68, 64–72. [Google Scholar]
- Day, M.; Zhao, X.; Wu, S.; Swanson, P.; Humphrey, P. Phorbol ester-induced apoptosis is accompanied by NGFI-A and c-fos activation in androgen-sensitive prostate cancer cells. Cell Growth Differ 1994, 5, 735–741. [Google Scholar]
- Rennecke, J.; Richter, K.H.; Haussermann, S.; Stempka, L.; Strand, S.; Stohr, M.; Marks, F. Biphasic effect of protein kinase C activators on spontaneous and glucocorticoid-induced apoptosis in primary mouse thymocytes. Biochim. Biophys. Acta Mol. Cell. Res 2000, 1497, 289–296. [Google Scholar]
- Hilario, M.C.; Puga, C.D.; Ocana, A.N.; Romo, F.P.G. Antioxidant activity, bioactive polyphenols in Mexican goats’ milk cheeses on summer grazing. J. Dairy Res 2010, 77, 20–26. [Google Scholar]
- Fujii, T.; Bermejo, M.L.G.; Bernabo, J.L.; Caamano, J.; Ohba, M.; Kuroki, T.; Li, L.; Yuspa, S.H.; Kazanietz, M.G. Involvement of protein kinase C (PKC) in phorbol ester-induced apoptosis in LNCaP prostate cancer cells. J. Biol. Chem 2000, 275, 7574–7582. [Google Scholar]
- Park, I.C.; Park, M.J.; Rhee, C.H.; Lee, J.I.; Choe, T.B.; Jang, J.J.; Lee, S.H.; Hong, S.I. Protein kinase C activation by PMA rapidly induces apoptosis through caspase-3/CPP32 and serine protease(s) in a gastric cancer cell line. Int. J. Oncol 2001, 18, 1077–1083. [Google Scholar]
- GraphPad Prism, version 5.01; GraphPad Software Inc.: San Diego, CA, USA, 2007.
- Association of Official Analytical Chemists (AOAC), Official Methods of Analysis, 17th ed; AOAC: Washington, DC, USA, 1990; pp. 64–87.
- Oskoueian, E.; Abdullah, N.; Ahmad, S.; Saad, W.Z.; Omar, A.R.; Ho, Y.W. Bioactive compounds and biological activities of Jatropha curcas L. kernel meal extract. Int. J. Mol. Sci 2011, 12, 5955–5970. [Google Scholar]
- Sharif, R.; Ghazali, A.; Rajab, N.; Haron, H.; Osman, F. Toxicological evaluation of some Malaysian locally processed raw food products. Food Chem. Toxicol 2008, 46, 368–374. [Google Scholar]
- Liu, B.; Li, C.Y.; Bian, H.J.; Min, M.W.; Chen, L.F.; Bao, J.K. Antiproliferative activity and apoptosis-inducing mechanism of Concanavalin A on human melanoma A375 cells. Arch. Biochem. Biophys 2009, 482, 1–6. [Google Scholar]
- Darzynkiewicz, Z.; Juan, G. Selective Extraction of Fragmented DNA from Apoptotic Cells for Analysis by Gel Electrophoresis and Identification of Apoptotic Cells by Flow Cytometry. In DNA-Repair Protocols, Eukaryotic Systems; Henderson, D.S., Ed.; Humana Press: New York, NY, USA, 1999; Volume 113, pp. 599–602. [Google Scholar]
- SAS, Statistical Analysis Institute, Version 9.1.3; SAS Institute Inc: Cary, NC, USA, 2003.

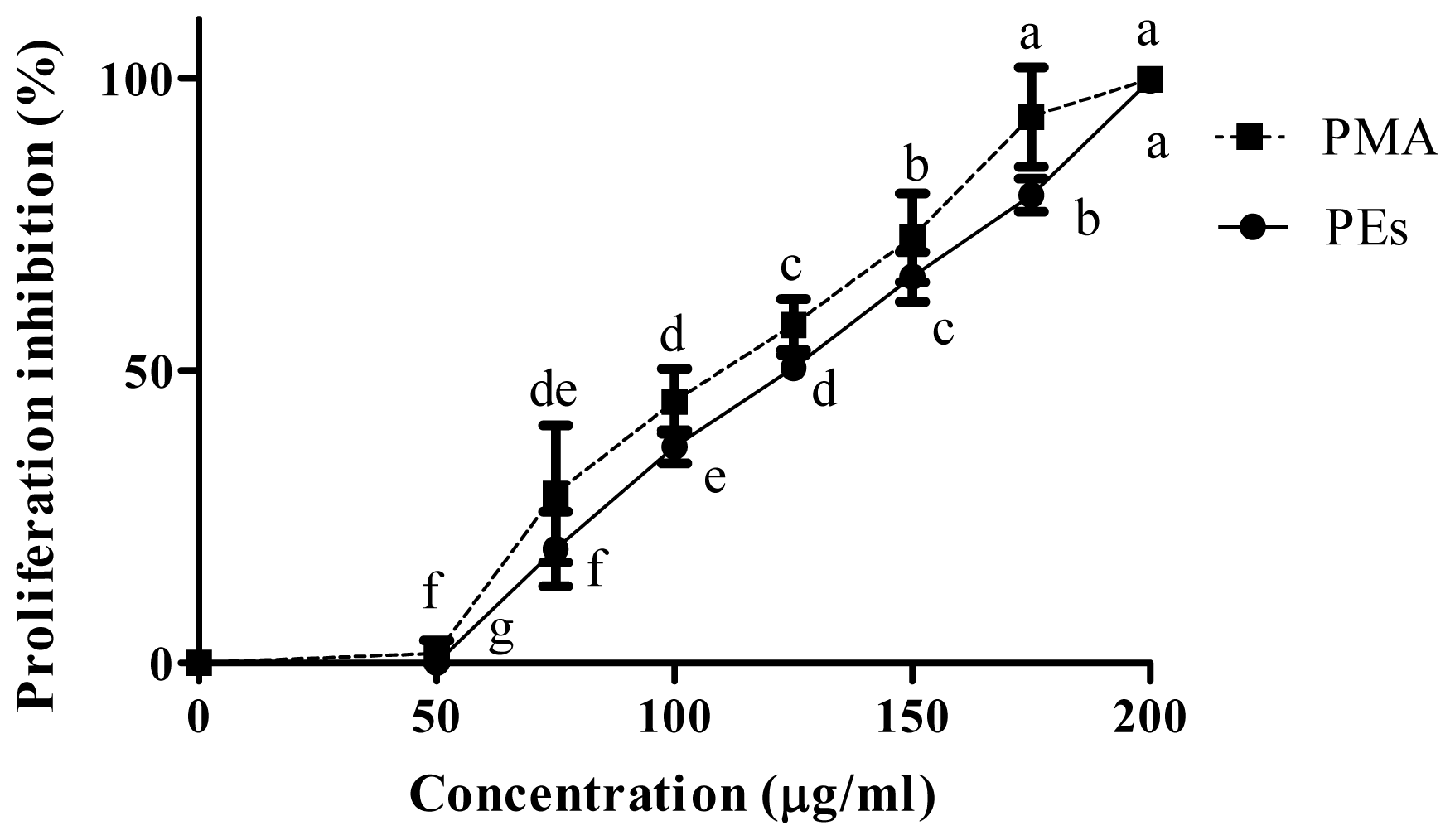
| CC50 μg/mL | |||
|---|---|---|---|
| Chang | Vero | S.E.M | |
| Isolated PEs | 125.9 a | 110.3 b | 2.79 |
| PMA1 | 124.5 a | 106.3 b | 3.06 |
| Chang Cells (%) | Vero Cells (%) | ||||||
|---|---|---|---|---|---|---|---|
| Un-treated | PEs-treated | PMA-treated | Un-treated | PEs-treated | PMA-treated | S.E.M | |
| Viable | 97.1 a | 32.3 b | 23.3 c | 97.0 a | 24.6 c | 17.3 d | 2.78 |
| Apoptotic | 1.6 d | 22.2 c | 27.6 b | 2.0 d | 29.3 b | 35.6 a | 2.28 |
| Dead | 2.0 b | 48.3 a | 51.6 a | 1.3 b | 50.6 a | 53.3 a | 3.47 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Oskoueian, E.; Abdullah, N.; Ahmad, S. Phorbol Esters Isolated from Jatropha Meal Induced Apoptosis-Mediated Inhibition in Proliferation of Chang and Vero Cell Lines. Int. J. Mol. Sci. 2012, 13, 13816-13829. https://doi.org/10.3390/ijms131113816
Oskoueian E, Abdullah N, Ahmad S. Phorbol Esters Isolated from Jatropha Meal Induced Apoptosis-Mediated Inhibition in Proliferation of Chang and Vero Cell Lines. International Journal of Molecular Sciences. 2012; 13(11):13816-13829. https://doi.org/10.3390/ijms131113816
Chicago/Turabian StyleOskoueian, Ehsan, Norhani Abdullah, and Syahida Ahmad. 2012. "Phorbol Esters Isolated from Jatropha Meal Induced Apoptosis-Mediated Inhibition in Proliferation of Chang and Vero Cell Lines" International Journal of Molecular Sciences 13, no. 11: 13816-13829. https://doi.org/10.3390/ijms131113816




