Superoxide Dismutase as a Novel Macromolecular Nitric Oxide Carrier: Preparation and Characterization
Abstract
:1. Introduction
2. Results and Discussion
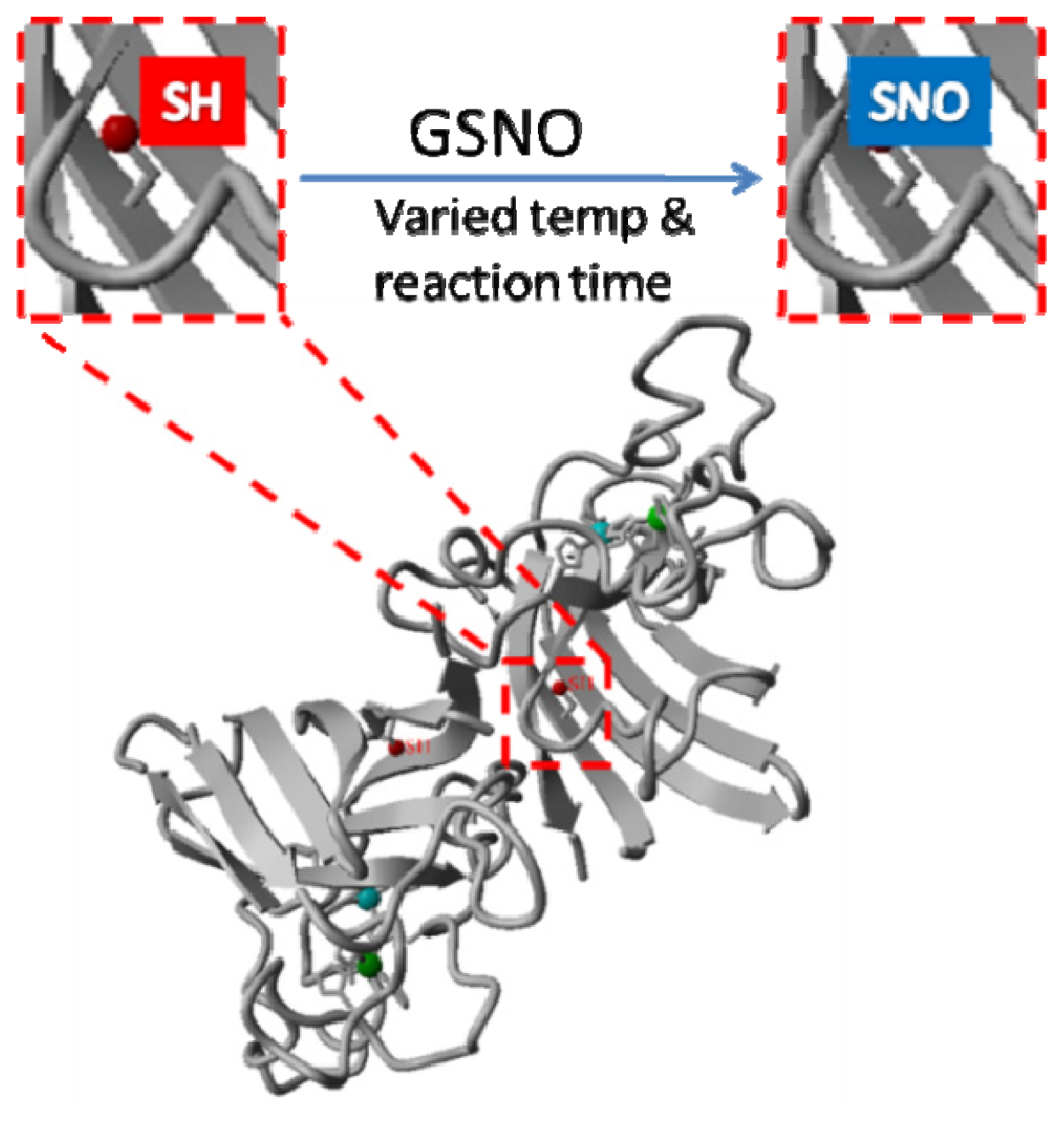
2.1. Rationale and Synthetic Protocol for SNO-bSOD
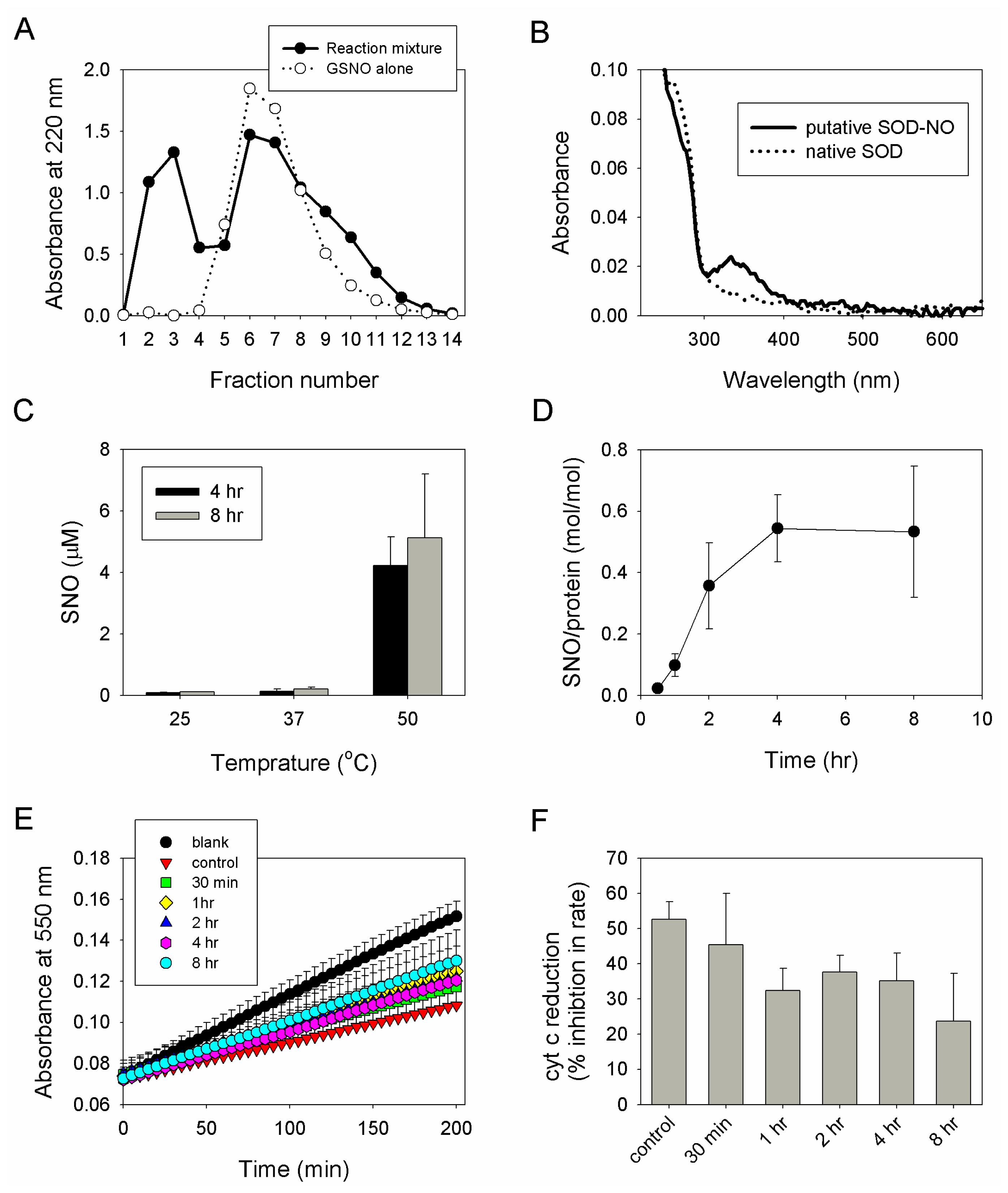
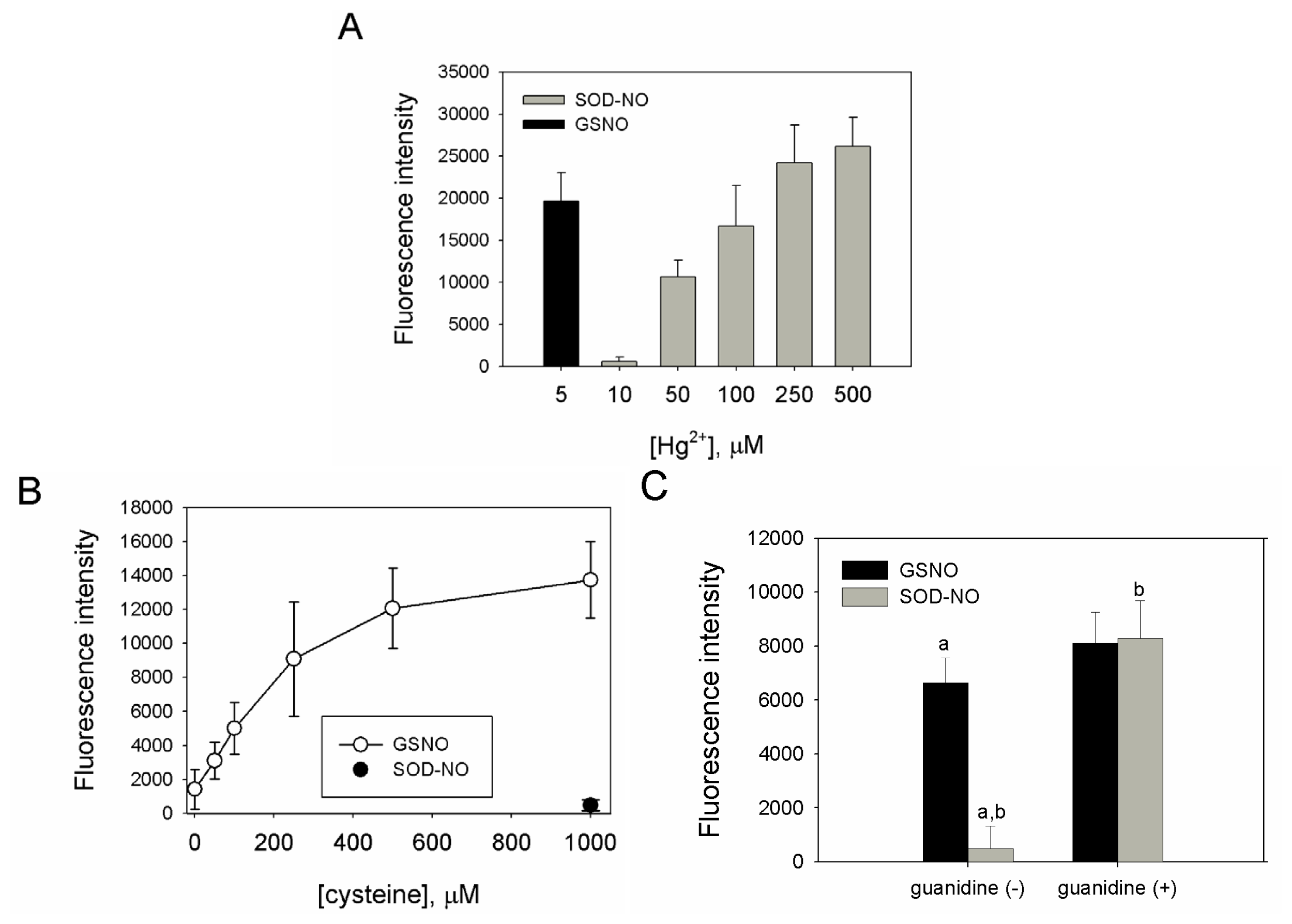
2.2. Preparation, Separation and Basic Characterization of SNO-bSOD
2.3. SNO-bSOD Is a Stable NO Carrier
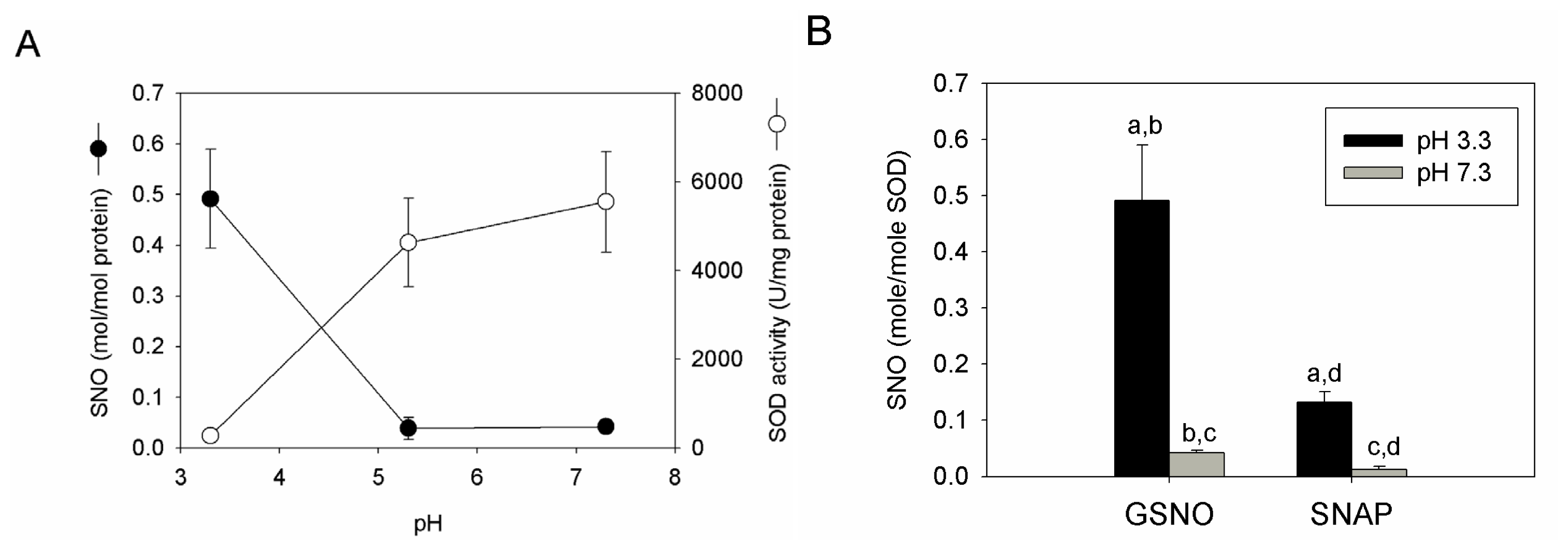
2.4. S-nitrosation Efficiency of bSOD Is Facilitated at Mildly Acidic pH
2.5. Discussion
3. Experimental Section
3.1. Chemicals
3.2. Synthesis of SNO-bSOD
3.3. Spectroscopic Characterization of SNO-bSOD
3.4. Fluorometric Determination of S-nitrosation Efficiency
3.5. Determination of Protein Concentration
3.6. Measurement of SOD Activity
3.7. Characterization of NO Release at Neutral pH
3.8. Quantification of Free Cysteine
3.9. Data Analysis
4. Conclusions
Acknowledgments
References
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: structure, function and inhibition. Biochem. J 2001, 357, 593–615. [Google Scholar]
- Wink, D.A.; Mitchell, J.B. Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic. Biol. Med 1998, 25, 434–456. [Google Scholar]
- Beckman, J.S.; Koppenol, W.H. Nitric oxide, superoxide, and peroxynitrite: The good, the bad, and ugly. Am. J. Physiol 1996, 271, C1424–C1437. [Google Scholar]
- Broniowska, K.A.; Hogg, N. The chemical biology of S-nitrosothiols. Antioxid. Redox. Signal 2012, 17, 969–980. [Google Scholar]
- Thomas, D.D.; Jourd’heuil, D. S-nitrosation: Current concepts and new developments. Antioxid. Redox. Signal 2012, 17, 934–936. [Google Scholar]
- Stamler, J.S.; Jaraki, O.; Osborne, J.; Simon, D.I.; Keaney, J.; Vita, J.; Singel, D.; Valeri, C.R.; Loscalzo, J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc. Natl. Acad. Sci. USA 1992, 89, 7674–7677. [Google Scholar]
- Scharfstein, J.S.; Keaney, J.F., Jr; Slivka, A.; Welch, G.N.; Vita, J.A.; Stamler, J.S.; Loscalzo, J. In vivotransfer of nitric oxide between a plasma protein-bound reservoir and low molecular weight thiols. J. Clin. Invest 1994, 94, 1432–1439. [Google Scholar]
- Jourd’heuil, D.; Hallen, K.; Feelisch, M.; Grisham, M.B. Dynamic state of S-nitrosothiols in human plasma and whole blood. Free Radic. Biol. Med 2000, 28, 409–417. [Google Scholar]
- Paige, J.S.; Xu, G.; Stancevic, B.; Jaffrey, S.R. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol 2008, 15, 1307–1316. [Google Scholar]
- Doulias, P.T.; Greene, J.L.; Greco, T.M.; Tenopoulou, M.; Seeholzer, S.H.; Dunbrack, R.L.; Ischiropoulos, H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc. Natl. Acad. Sci. USA 2010, 107, 16958–16963. [Google Scholar]
- Seth, D.; Stamler, J.S. The SNO-proteome: Causation and classifications. Curr. Opin. Chem. Biol 2011, 15, 129–136. [Google Scholar]
- Foster, M.W. Methodologies for the characterization, identification and quantification of S-nitrosylated proteins. Biochim. Biophys. Acta 2011, 1820, 675–683. [Google Scholar]
- Chen, Y.J.; Ku, W.C.; Lin, P.Y.; Chou, H.C.; Khoo, K.H.; Chen, Y.J. S-alkylating labeling strategy for site-specific identification of the s-nitrosoproteome. J. Proteome Res 2010, 9, 6417–6439. [Google Scholar]
- Stamler, J.S.; Simon, D.I.; Osborne, J.A.; Mullins, M.E.; Jaraki, O.; Michel, T.; Singel, D.J.; Loscalzo, J. S-nitrosylation of proteins with nitric oxide: Synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. USA 1992, 89, 444–448. [Google Scholar]
- Marks, D.S.; Vita, J.A.; Folts, J.D.; Keaney, J.F., Jr; Welch, G.N.; Loscalzo, J. Inhibition of neointimal proliferation in rabbits after vascular injury by a single treatment with a protein adduct of nitric oxide. J. Clin. Invest 1995, 96, 2630–2638. [Google Scholar]
- Ewing, J.F.; Young, D.V.; Janero, D.R.; Garvey, D.S.; Grinnell, T.A. Nitrosylated bovine serum albumin derivatives as pharmacologically active nitric oxide congeners. J. Pharmacol. Exp. Ther 1997, 283, 947–954. [Google Scholar]
- Katsumi, H.; Nishikawa, M.; Ma, S.F.; Yamashita, F.; Hashida, M. Physicochemical, tissue distribution, and vasodilation characteristics of nitrosated serum albumin: delivery of nitric oxide in vivo. J. Pharm. Sci 2004, 93, 2343–2352. [Google Scholar]
- Katsumi, H.; Nishikawa, M.; Yamashita, F.; Hashida, M. Development of polyethylene glycol-conjugated poly-S-nitrosated serum albumin, a novel S-Nitrosothiol for prolonged delivery of nitric oxide in the blood circulation in vivo. J. Pharmacol. Exp. Ther 2005, 314, 1117–1124. [Google Scholar]
- Ishima, Y.; Hiroyama, S.; Kragh-Hansen, U.; Maruyama, T.; Sawa, T.; Akaike, T.; Kai, T.; Otagiri, M. One-step preparation of S-nitrosated human serum albumin with high biological activities. Nitric Oxide 2010, 23, 121–127. [Google Scholar]
- Ishima, Y.; Yoshida, F.; Kragh-Hansen, U.; Watanabe, K.; Katayama, N.; Nakajou, K.; Akaike, T.; Kai, T.; Maruyama, T.; Otagiri, M. Cellular uptake mechanisms and responses to NO transferred from mono- and poly-S-nitrosated human serum albumin. Free Radic. Res 2011, 45, 1196–1206. [Google Scholar]
- Ishima, Y.; Akaike, T.; Kragh-Hansen, U.; Hiroyama, S.; Sawa, T.; Suenaga, A.; Maruyama, T.; Kai, T.; Otagiri, M. S-nitrosylated human serum albumin-mediated cytoprotective activity is enhanced by fatty acid binding. J. Biol. Chem 2008, 283, 34966–34975. [Google Scholar]
- Ishima, Y.; Kragh-Hansen, U.; Maruyama, T.; Otagiri, M. Albumin as a nitric oxide-traffic protein: Characterization, biochemistry and possible future therapeutic applications. Drug Metab. Pharmacokinet 2009, 24, 308–317. [Google Scholar]
- Katayama, N.; Nakajou, K.; Komori, H.; Uchida, K.; Yokoe, J.; Yasui, N.; Yamamoto, H.; Kai, T.; Sato, M.; Nakagawa, T.; et al. Design and evaluation of S-nitrosylated human serum albumin as a novel anticancer drug. J. Pharmacol. Exp. Ther 2008, 325, 69–76. [Google Scholar]
- Katayama, N.; Nakajou, K.; Ishima, Y.; Ikuta, S.; Yokoe, J.; Yoshida, F.; Suenaga, A.; Maruyama, T.; Kai, T.; Otagiri, M. Nitrosylated human serum albumin (SNO-HSA) induces apoptosis in tumor cells. Nitric Oxide 2010, 22, 259–265. [Google Scholar]
- Ishima, Y.; Chen, D.; Fang, J.; Maeda, H.; Minomo, A.; Kragh-Hansen, U.; Kai, T.; Maruyama, T.; Otagiri, M. S-Nitrosated human serum albumin dimer is not only a novel anti-tumor drug but also a potentiator for anti-tumor drugs with augmented EPR effects. Bioconjug. Chem 2012, 23, 264–271. [Google Scholar]
- McCord, J.M.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem 1969, 244, 6049–6055. [Google Scholar]
- Rypniewski, W.R.; Mangani, S.; Bruni, B.; Orioli, P.L.; Casati, M.; Wilson, K.S. Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution. J. Mol. Biol 1995, 251, 282–296. [Google Scholar]
- Carden, D.L.; Granger, D.N. Pathophysiology of ischaemia-reperfusion injury. J. Pathol 2000, 190, 255–266. [Google Scholar]
- Kurose, I.; Wolf, R.; Grisham, M.B.; Granger, D.N. Modulation of ischemia/reperfusion-induced microvascular dysfunction by nitric oxide. Circ. Res 1994, 74, 376–382. [Google Scholar]
- Ferdinandy, P.; Schulz, R. Nitric oxide, superoxide, and peroxynitrite in myocardial ischaemia-reperfusion injury and preconditioning. Br. J. Pharmacol 2003, 138, 532–543. [Google Scholar]
- Jung, O.; Marklund, S.L.; Geiger, H.; Pedrazzini, T.; Busse, R.; Brandes, R.P. Extracellular superoxide dismutase is a major determinant of nitric oxide bioavailability: In vivo and ex vivo evidence from ecSOD-deficient mice. Circ. Res 2003, 93, 622–629. [Google Scholar]
- Katsumi, H.; Nishikawa, M.; Yasui, H.; Yamashita, F.; Hashida, M. Prevention of ischemia/reperfusion injury by hepatic targeting of nitric oxide in mice. J. Control. Release 2009, 140, 12–17. [Google Scholar]
- Konorev, E.A.; Kalyanaraman, B.; Hogg, N. Modification of creatine kinase by S-nitrosothiols: S-nitrosation vs. S-thiolation. Free Radic. Biol. Med 2000, 28, 1671–1678. [Google Scholar]
- Giustarini, D.; Milzani, A.; Aldini, G.; Carini, M.; Rossi, R.; Dalle-Donne, I. S-nitrosation versus S-glutathionylation of protein sulfhydryl groups by S-nitrosoglutathione. Antioxid. Redox. Signal 2005, 7, 930–939. [Google Scholar]
- Wolzt, M.; MacAllister, R.J.; Davis, D.; Feelisch, M.; Moncada, S.; Vallance, P.; Hobbs, A.J. Biochemical characterization of S-nitrosohemoglobin. Mechanisms underlying synthesis, no release, and biological activity. J. Biol. Chem 1999, 274, 28983–28990. [Google Scholar]
- Barglow, K.T.; Knutson, C.G.; Wishnok, J.S.; Tannenbaum, S.R.; Marletta, M.A. Site-specific and redox-controlled S-nitrosation of thioredoxin. Proc. Natl. Acad. Sci. USA 2011, 108, E600–E606. [Google Scholar]
- Singh, R.J.; Hogg, N.; Joseph, J.; Kalyanaraman, B. Mechanism of nitric oxide release from S-nitrosothiols. J. Biol. Chem 1996, 271, 18596–18603. [Google Scholar]
- Stamler, J.S.; Toone, E.J. The decomposition of thionitrites. Curr. Opin. Chem. Biol 2002, 6, 779–785. [Google Scholar]
- Gorren, A.C.; Schrammel, A.; Schmidt, K.; Mayer, B. Decomposition of S-nitrosoglutathione in the presence of copper ions and glutathione. Arch. Biochem. Biophys 1996, 330, 219–228. [Google Scholar]
- Arnelle, D.R.; Stamler, J.S. NO+, NO•, and NO− donation by S-nitrosothiols: Implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys 1995, 318, 279–285. [Google Scholar]
- Hu, T.M.; Chou, T.C. The kinetics of thiol-mediated decomposition of S-nitrosothiols. Aaps. J 2006, 8, E485–E492. [Google Scholar]
- Meyer, D.J.; Kramer, H.; Ozer, N.; Coles, B.; Ketterer, B. Kinetics and equilibria of S-nitrosothiol-thiol exchange between glutathione, cysteine, penicillamines and serum albumin. FEBS Lett 1994, 345, 177–180. [Google Scholar]
- Cook, J.A.; Kim, S.Y.; Teague, D.; Krishna, M.C.; Pacelli, R.; Mitchell, J.B.; Vodovotz, Y.; Nims, R.W.; Christodoulou, D.; Miles, A.M.; et al. Convenient colorimetric and fluorometric assays for S-nitrosothiols. Anal. Biochem 1996, 238, 150–158. [Google Scholar]
- Scorza, G.; Pietraforte, D.; Minetti, M. Role of ascorbate and protein thiols in the release of nitric oxide from S-nitroso-albumin and S-nitroso-glutathione in human plasma. Free Radic. Biol. Med 1997, 22, 633–642. [Google Scholar]
- Hallewell, R.A.; Imlay, K.C.; Lee, P.; Fong, N.M.; Gallegos, C.; Getzoff, E.D.; Tainer, J.A.; Cabelli, D.E.; Tekamp-Olson, P.; Mullenbach, G.T.; et al. Thermostabilization of recombinant human and bovine CuZn superoxide dismutases by replacement of free cysteines. Biochem. Biophys. Res. Commun 1991, 181, 474–480. [Google Scholar]
- Lepock, J.R.; Frey, H.E.; Hallewell, R.A. Contribution of conformational stability and reversibility of unfolding to the increased thermostability of human and bovine superoxide dismutase mutated at free cysteines. J. Biol. Chem 1990, 265, 21612–21618. [Google Scholar]
- Fee, J.A.; Phillips, W.D. The behavior of holo- and apo-forms of bovine superoxide dismutase at low pH. Biochim. Biophys. Acta 1975, 412, 26–38. [Google Scholar]
- Padgett, C.M.; Whorton, A.R. Cellular responses to nitric oxide: Role of protein S-thiolation/dethiolation. Arch. Biochem. Biophys 1998, 358, 232–242. [Google Scholar]
- Ji, Y.; Akerboom, T.P.; Sies, H.; Thomas, J.A. S-nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch. Biochem. Biophys 1999, 362, 67–78. [Google Scholar]
- Mohr, S.; Hallak, H.; de Boitte, A.; Lapetina, E.G.; Brune, B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem 1999, 274, 9427–9430. [Google Scholar]
- Li, J.; Huang, F.L.; Huang, K.P. Glutathiolation of proteins by glutathione disulfide S-oxide derived from S-nitrosoglutathione. Modifications of rat brain neurogranin/RC3 and neuromodulin/GAP-43. J. Biol. Chem 2001, 276, 3098–3105. [Google Scholar]
- Xian, M.; Chen, X.; Liu, Z.; Wang, K.; Wang, P.G. Inhibition of papain by S-nitrosothiols. Formation of mixed disulfides. J. Biol. Chem 2000, 275, 20467–20473. [Google Scholar]
- Liu, Z.; Rudd, M.A.; Freedman, J.E.; Loscalzo, J. S-Transnitrosation reactions are involved in the metabolic fate and biological actions of nitric oxide. J. Pharmacol. Exp. Ther 1998, 284, 526–534. [Google Scholar]
- Hogg, N. The kinetics of S-transnitrosation—A reversible second-order reaction. Anal. Biochem 1999, 272, 257–262. [Google Scholar]
- Klatt, P.; Lamas, S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur. J. Biochem 2000, 267, 4928–4944. [Google Scholar]
- Beckman, J.S.; Minor, R.L., Jr; White, C.W.; Repine, J.E.; Rosen, G.M.; Freeman, B.A. Superoxide dismutase and catalase conjugated to polyethylene glycol increases endothelial enzyme activity and oxidant resistance. J. Biol. Chem. 1988, 263, 6884–6892. [Google Scholar]
- Filipe, P.; Emerit, I.; Vassy, J.; Levy, A.; Huang, V.; Freitas, J. Cellular penetration of fluorescently labeled superoxide dismutases of various origins. Mol. Med 1999, 5, 517–525. [Google Scholar]
- Chorny, M.; Hood, E.; Levy, R.J.; Muzykantov, V.R. Endothelial delivery of antioxidant enzymes loaded into non-polymeric magnetic nanoparticles. J. Control. Release 2010, 146, 144–151. [Google Scholar]
- Hood, E.; Simone, E.; Wattamwar, P.; Dziubla, T.; Muzykantov, V. Nanocarriers for vascular delivery of antioxidants. Nanomedicine (Lond) 2011, 6, 1257–1272. [Google Scholar]
- Yamakura, F.; Taka, H.; Fujimura, T.; Murayama, K. Inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J. Biol. Chem 1998, 273, 14085–14089. [Google Scholar]
- Demicheli, V.; Quijano, C.; Alvarez, B.; Radi, R. Inactivation and nitration of human superoxide dismutase (SOD) by fluxes of nitric oxide and superoxide. Free Radic. Biol. Med 2007, 42, 1359–1368. [Google Scholar]
- Akhtar, M.W.; Sunico, C.R.; Nakamura, T.; Lipton, S.A. Redox Regulation of Protein Function via Cysteine S-Nitrosylation and Its Relevance to Neurodegenerative Diseases. Int. J. Cell Biol. 2012. [Google Scholar] [CrossRef]
- Brown, G.C. Nitric oxide and neuronal death. Nitric Oxide 2010, 23, 153–165. [Google Scholar]
- Calabrese, V.; Cornelius, C.; Rizzarelli, E.; Owen, J.B.; Dinkova-Kostova, A.T.; Butterfield, D.A. Nitric oxide in cell survival: A janus molecule. Antioxid. Redox. Signal 2009, 11, 2717–2739. [Google Scholar]
- Steinert, J.R.; Chernova, T.; Forsythe, I.D. Nitric oxide signaling in brain function, dysfunction, and dementia. Neuroscientist 2010, 16, 435–452. [Google Scholar]
- Uehara, T.; Nakamura, T.; Yao, D.; Shi, Z.Q.; Gu, Z.; Ma, Y.; Masliah, E.; Nomura, Y.; Lipton, S.A. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 2006, 441, 513–517. [Google Scholar]
- Fujiwara, N.; Nakano, M.; Kato, S.; Yoshihara, D.; Ookawara, T.; Eguchi, H.; Taniguchi, N.; Suzuki, K. Oxidative modification to cysteine sulfonic acid of Cys111 in human copper-zinc superoxide dismutase. J. Biol. Chem 2007, 282, 35933–35944. [Google Scholar]
- Okado-Matsumoto, A.; Guan, Z.; Fridovich, I. Modification of cysteine 111 in human Cu,Zn-superoxide dismutase. Free Radic. Biol. Med 2006, 41, 1837–1846. [Google Scholar]
- Stathopulos, P.B.; Rumfeldt, J.A.; Scholz, G.A.; Irani, R.A.; Frey, H.E.; Hallewell, R.A.; Lepock, J.R.; Meiering, E.M. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 7021–7026. [Google Scholar]
- Rumfeldt, J.A.; Stathopulos, P.B.; Chakrabarrty, A.; Lepock, J.R.; Meiering, E.M. Mechanism and thermodynamics of guanidinium chloride-induced denaturation of ALS-associated mutant Cu,Zn superoxide dismutases. J. Mol. Biol 2006, 355, 106–123. [Google Scholar]
- Rumfeldt, J.A.; Lepock, J.R.; Meiering, E.M. Unfolding and folding kinetics of amyotrophic lateral sclerosis-associated mutant Cu,Zn superoxide dismutases. J. Mol. Biol 2009, 385, 278–298. [Google Scholar]
- Antharavally, B.S.; Mallia, K.A.; Rangaraj, P.; Haney, P.; Bell, P.A. Quantitation of proteins using a dye-metal-based colorimetric protein assay. Anal. Biochem 2009, 385, 342–345. [Google Scholar]
- Abernethy, J.L.; Steinman, H.M.; Hill, R.L. Bovine erythrocyte superoxide dismutase. Subunit structure and sequence location of the intrasubunit disulfide bond. J. Biol. Chem 1974, 249, 7339–7347. [Google Scholar]

© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chen, S.-H.; Chiu, S.-J.; Hu, T.-M. Superoxide Dismutase as a Novel Macromolecular Nitric Oxide Carrier: Preparation and Characterization. Int. J. Mol. Sci. 2012, 13, 13985-14001. https://doi.org/10.3390/ijms131113985
Chen S-H, Chiu S-J, Hu T-M. Superoxide Dismutase as a Novel Macromolecular Nitric Oxide Carrier: Preparation and Characterization. International Journal of Molecular Sciences. 2012; 13(11):13985-14001. https://doi.org/10.3390/ijms131113985
Chicago/Turabian StyleChen, Ssu-Han, Shih-Jiuan Chiu, and Teh-Min Hu. 2012. "Superoxide Dismutase as a Novel Macromolecular Nitric Oxide Carrier: Preparation and Characterization" International Journal of Molecular Sciences 13, no. 11: 13985-14001. https://doi.org/10.3390/ijms131113985



