The Murine PSE/TATA-Dependent Transcriptome: Evidence of Functional Homologies with Its Human Counterpart
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Screening in silico of the Mouse Genome Discloses a Large Number of Putative snRNA-Like Transcriptional Units That Map Preferentially in Intronic Regions of Protein-Coding Genes
2.2. Significant Association to “Alternative Splicing” and “Brain” Functional Annotations of Coding Genes Hosting Pol III Transcriptional Units
2.3. 38B ncRNA Is the Murine Functional Homolog of the Human 38A ncRNA
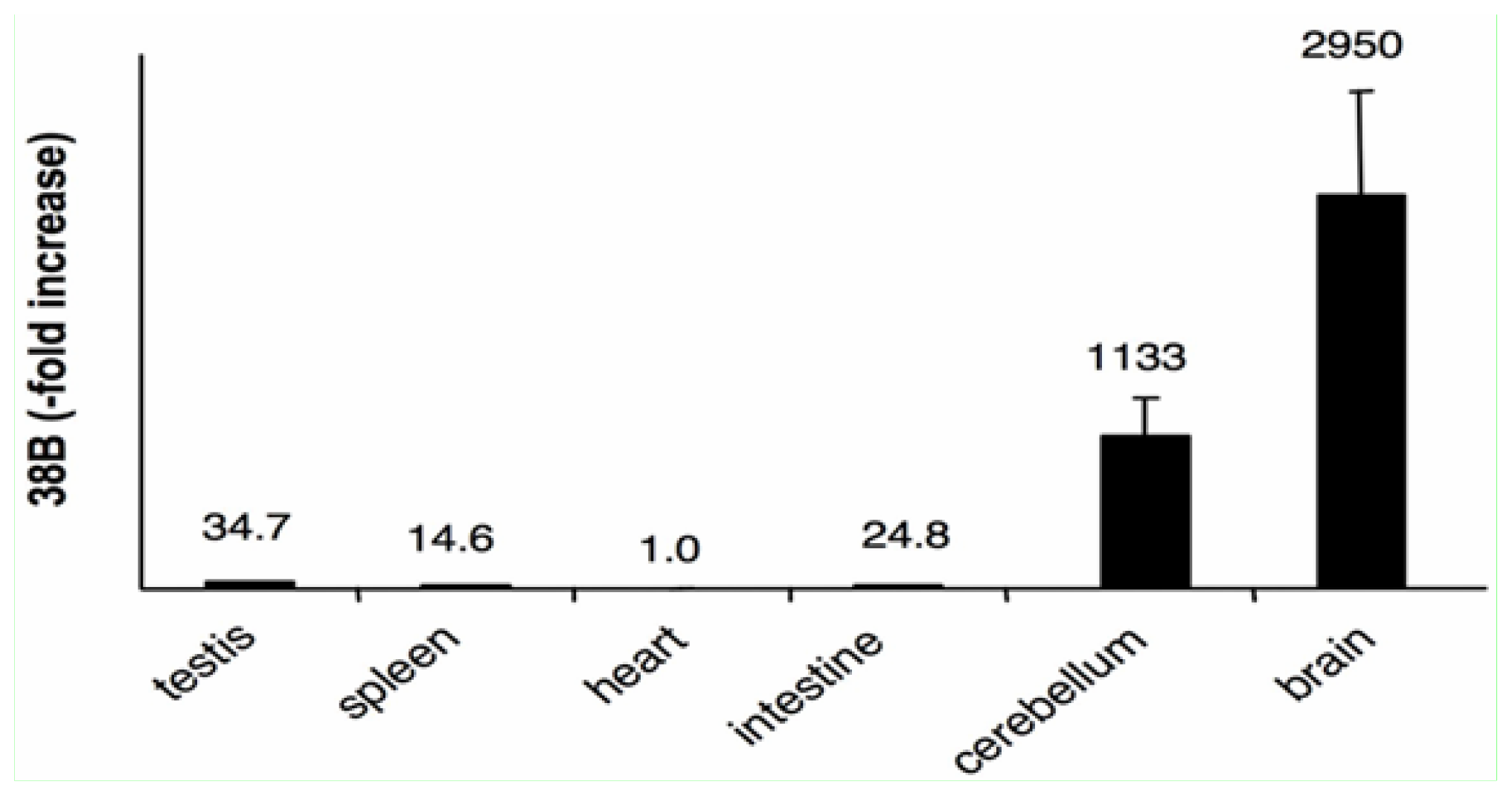
2.4. The Expression of 38B RNA Is Specifically Associated to the Brain
2.5. 38B Expression Leads to the Synthesis of KCNIP4 Alternative Variant C in Mouse
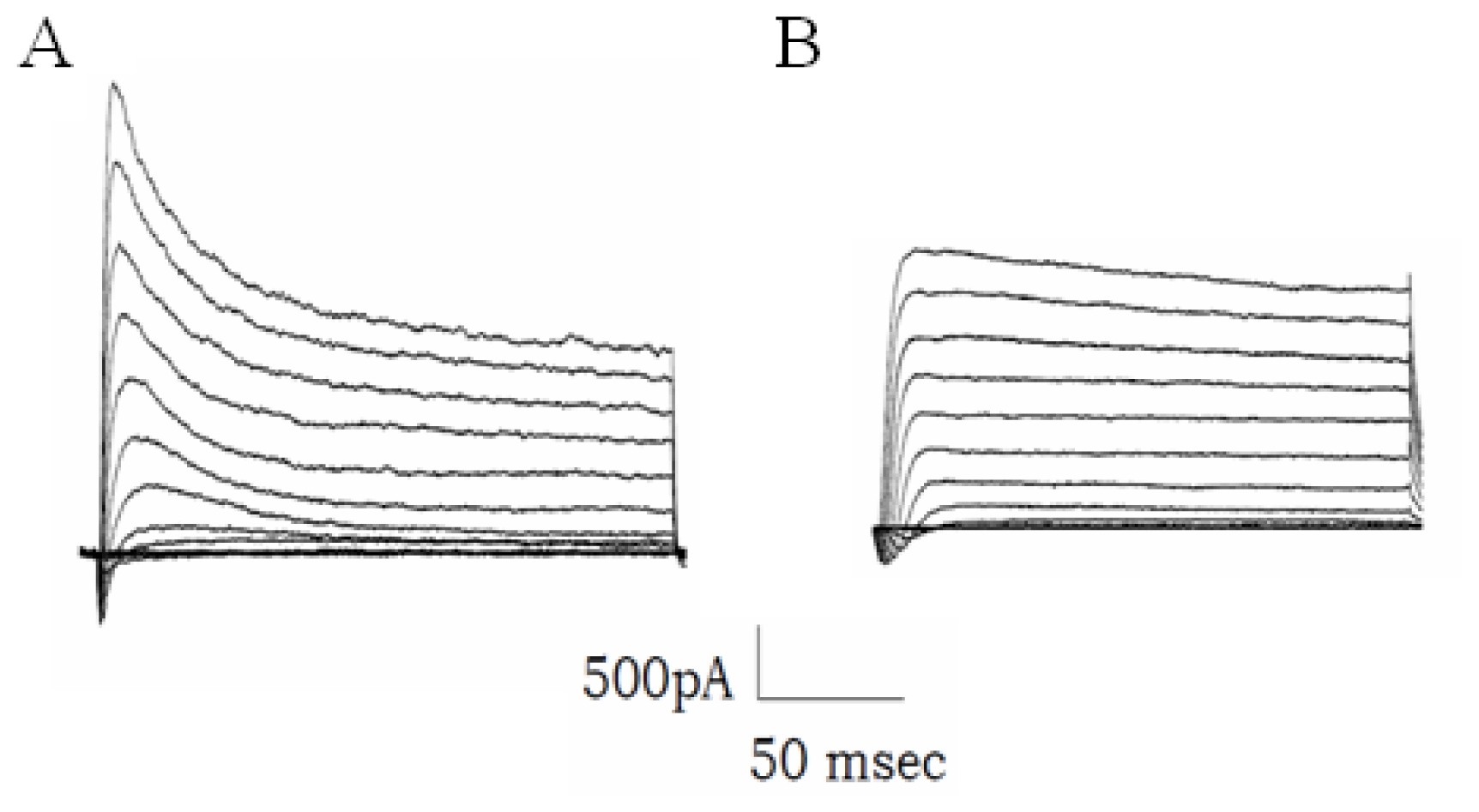
2.6. The Increased Transcription of 38B Affects Fast Inactivation of Mouse A-Type K+ Channels
3. Experimental Section
3.1. Screening of the Mouse Genome with COMPASSS and Identification of Pol III Type 3 Transcriptional Units
3.2. Statistical Analysis
3.3. Gene Annotations
3.4. 38B Plasmid Constructs Generation
3.5. In vitro Transcription
3.6. Hippocampal Neurons Preparation and Transfection
3.7. Real Time Quantitative RT PCR Analysis
3.8. Electrophysiology
4. Conclusions
Supplementary Materials
ijms-13-14813-s001.pdfAcknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev 2002, 16, 2593–2620. [Google Scholar]
- Pagano, A.; Castelnuovo, M.; Tortelli, F.; Ferrari, R.; Dieci, G.; Cancedda, R. New small nuclear RNA gene-like transcriptional units as sources of regulatory transcripts. PLoS Genet 2007, 3, e1. [Google Scholar]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet 2007, 23, 614–622. [Google Scholar]
- Orioli, A.; Pascali, C.; Pagano, A.; Teichmann, M.; Dieci, G. RNA polymerase III transcription control elements: Themes and variations. Gene 2012, 493, 185–194. [Google Scholar]
- Castelnuovo, M.; Massone, S.; Tasso, R.; Fiorino, G.; Gatti, M.; Robello, M.; Gatta, E.; Berger, A.; Strub, K.; Florio, T.; et al. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J 2010, 24, 4033–4046. [Google Scholar]
- Massone, S.; Vassallo, I.; Castelnuovo, M.; Fiorino, G.; Gatta, E.; Robello, M.; Borghi, R.; Tabaton, M.; Russo, C.; Dieci, G.; et al. RNA polymerase III drives alternative splicing of the potassium channel-interacting protein contributing to brain complexity and neurodegeneration. J. Cell Biol 2011, 193, 851–866. [Google Scholar]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis 2011, 41, 308–317. [Google Scholar]
- Barski, A.; Chepelev, I.; Liko, D.; Cuddapah, S.; Fleming, A.B.; Birch, J.; Cui, K.; White, R.J.; Zhao, K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol 2012, 17, 629–634. [Google Scholar]
- Canella, D.; Praz, V.; Reina, J.H.; Cousin, P.; Hernandez, N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res 2010, 20, 710–721. [Google Scholar]
- Oler, A.J.; Alla, R.K.; Roberts, D.N.; Wong, A.; Hollenhorst, P.C.; Chandler, K.J.; Cassiday, P.A.; Nelson, C.A.; Hagedorn, C.H.; Graves, B.J.; et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol 2012, 17, 620–628. [Google Scholar]
- Carrière, L.; Graziani, S.; Alibert, O.; Ghavi-Helm, Y.; Boussouar, F.; Humbertclaude, H.; Jounier, S.; Aude, J.C.; Keime, C.; Murvai, J.; et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res 2012, 40, 270–283. [Google Scholar]
- Dieci, G.; Conti, A.; Pagano, A.; Carnevali, D. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta 2012, in press. [Google Scholar]
- Maccari, G.; Gemignani, F.; Landi, S. COMPASSS (COMplex PAttern of Sequence Search Software), a simple and effective tool for mining complex motifs in whole genomes. Bioinformatics 2010, 26, 1777–1778. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009, 4, 57. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009, 37, 1–13. [Google Scholar]
- Garritano, S.; Gigoni, A.; Costa, D.; Malatesta, P.; Florio, T.; Cancedda, R.; Pagano, A. A Novel Collection of snRNA-like promoters with Tissue-Specific Transcription Properties. Int. J. Mol. Sci 2012, 13, 11323–11331. [Google Scholar]
- Orioli, A.; Pascali, C.; Quartararo, J.; Diebel, K.W.; Praz, V.; Romascano, D.; Percudani, R.; van Dyk, L.F.; Hernandez, N.; Teichmann, M.; et al. Widespread occurrence of non-canonical transcription termination by human RNA polymerase III. Nucleic Acids Res 2012, 39, 5499–5512. [Google Scholar]
- Baranauskas, G. Cell-type-specific splicing of KChIP4 mRNA correlates with slower kinetics of A-type current. Eur. J. Neurosci 2004, 20, 385–391. [Google Scholar]
- Holmqvist, M.H.; Cao, J.; Hernandez-Pineda, R.; Jacobson, M.D.; Carroll, K.I.; Sung, M.A.; Betty, M.; Ge, P.; Gilbride, K.J.; Brown, M.E.; et al. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc. Natl. Acad. Sci. USA 2002, 99, 1035–1040. [Google Scholar]
- Pruunsild, P.; Timmusk, T. Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics 2005, 86, 581–593. [Google Scholar]
- Angulo, E.; Noé, V.; Casadó, V.; Mallol, J.; Gomez-Isla, T.; Lluis, C.; Ferrer, I.; Ciudad, C.J.; Franco, R. Up-regulation of the Kv3.4 potassium channel subunit in early stages of Alzheimer’s disease. J. Neurochem 2004, 91, 547–557. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Pagano, A.; Giannoni, P.; Zambotti, A.; Randazzo, N.; Zerega, B.; Cancedda, R.; Dozin, B. CALbeta, a novel lipocalin associated with chondrogenesis and inflammation. Eur. J. Cell Biol 2002, 81, 264–272. [Google Scholar]
- Zerega, B.; Pagano, A.; Pianezzi, A.; Ulivi, V.; Camardella, L.; Cancedda, R.; Cancedda, F.D. Expression of serum amyloid A in chondrocytes and myoblasts differentiation and inflammation: possible role in cholesterol homeostasis. Matrix Biol 2004, 23, 35–46. [Google Scholar]
- Gavazzo, P.; Vella, S.; Marchetti, C.; Nizzari, M.; Cancedda, R.; Pagano, A. Acquisition of neuron-like electrophysiological properties in neuroblastoma cells by controlled expression of NDM29 ncRNA. J. Neurochem 2012, 119, 989–1001. [Google Scholar]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar]
- Mattick, J.S. The hidden genetic program of complex organisms. Sci. Am 2004, 291, 60–67. [Google Scholar]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar]
- Mills, J.D.; Janitz, M. Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol. Aging 2012, 33, e11–e24. [Google Scholar]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet 2008, 40, 1413–1415. [Google Scholar]
- Tsirigos, A.; Rigoutsos, I. Human and mouse introns are linked to the same processes and functions through each genome’s most frequent non-conserved motifs. Nucleic Acids Res 2008, 36, 3484–3493. [Google Scholar]



| Category | Term | Count | % | p-Value | List Total | Pop Hits | Pop Total | Bonferroni | Benjamini |
|---|---|---|---|---|---|---|---|---|---|
| SP_PIR_KEYWORDS | alternative splicing | 227 | 44.77 | 4.64 × 10−29 | 464 | 4481 | 17854 | 1.48 × 10−26 | 1.48 × 10−26 |
| UP_SEQ_FEATURE | splice variant | 223 | 43.98 | 1.46 × 10−24 | 442 | 4448 | 16021 | 2.37 × 10−21 | 2.37 × 10−21 |
| UP_TISSUE | Brain | 273 | 53.85 | 1.44 × 10−17 | 493 | 7313 | 19868 | 2.28 × 10−15 | 2.28 × 10−15 |
| UP_SEQ_FEATURE | domain: EGF-like 1 | 21 | 4.14 | 8.94 × 10−12 | 442 | 106 | 16021 | 1.45 × 10−8 | 7.23 × 10−9 |
| SP_PIR_KEYWORDS | phosphoprotein | 229 | 45.17 | 3.63 × 10−10 | 464 | 6311 | 17854 | 1.15 × 10−7 | 5.77 × 10−8 |
| UP_SEQ_FEATURE | domain: EGF-like 2 | 17 | 3.35 | 3.65 × 10−10 | 442 | 79 | 16021 | 5.89 × 10−7 | 1.96 × 10−7 |
| GOTERM_BP_FAT | GO:0007155 ~cell adhesion | 43 | 8.48 | 5.00 × 10−10 | 352 | 561 | 13588 | 8.32 × 10−7 | 8.32 × 10−7 |
| GOTERM_BP_FAT | GO:0022610 ~biological adhesion | 43 | 8.48 | 5.30 × 10−10 | 352 | 562 | 13588 | 8.83 × 10−7 | 4.41 × 10−7 |
| SP_PIR_KEYWORDS | egf-like domain | 26 | 5.13 | 7.54 × 10−10 | 464 | 222 | 17854 | 2.40 × 10−7 | 7.99 × 10−8 |
| UP_TISSUE | Cerebellum | 80 | 15.78 | 1.33 × 10−9 | 493 | 1580 | 19868 | 2.10 × 10−7 | 1.05E × 10−7 |
| UP_SEQ_FEATURE | compositionally biased region: Ser-rich | 34 | 6.71 | 5.78 × 10−9 | 442 | 380 | 16021 | 9.35 × 10−6 | 2.34 × 10−6 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bruzzone, M.J.; Gavazzo, P.; Massone, S.; Balbi, C.; Villa, F.; Conti, A.; Dieci, G.; Cancedda, R.; Pagano, A. The Murine PSE/TATA-Dependent Transcriptome: Evidence of Functional Homologies with Its Human Counterpart. Int. J. Mol. Sci. 2012, 13, 14813-14827. https://doi.org/10.3390/ijms131114813
Bruzzone MJ, Gavazzo P, Massone S, Balbi C, Villa F, Conti A, Dieci G, Cancedda R, Pagano A. The Murine PSE/TATA-Dependent Transcriptome: Evidence of Functional Homologies with Its Human Counterpart. International Journal of Molecular Sciences. 2012; 13(11):14813-14827. https://doi.org/10.3390/ijms131114813
Chicago/Turabian StyleBruzzone, Maria Jessica, Paola Gavazzo, Sara Massone, Carolina Balbi, Federico Villa, Anastasia Conti, Giorgio Dieci, Ranieri Cancedda, and Aldo Pagano. 2012. "The Murine PSE/TATA-Dependent Transcriptome: Evidence of Functional Homologies with Its Human Counterpart" International Journal of Molecular Sciences 13, no. 11: 14813-14827. https://doi.org/10.3390/ijms131114813






