3.1. Nickel
NiCl
2 is not extremely toxic to
D. discoideum, probably because of an energy-dependent nickel export that exists in wild-type and mutant strains, but its role in heavy metal resistance has not been confirmed [
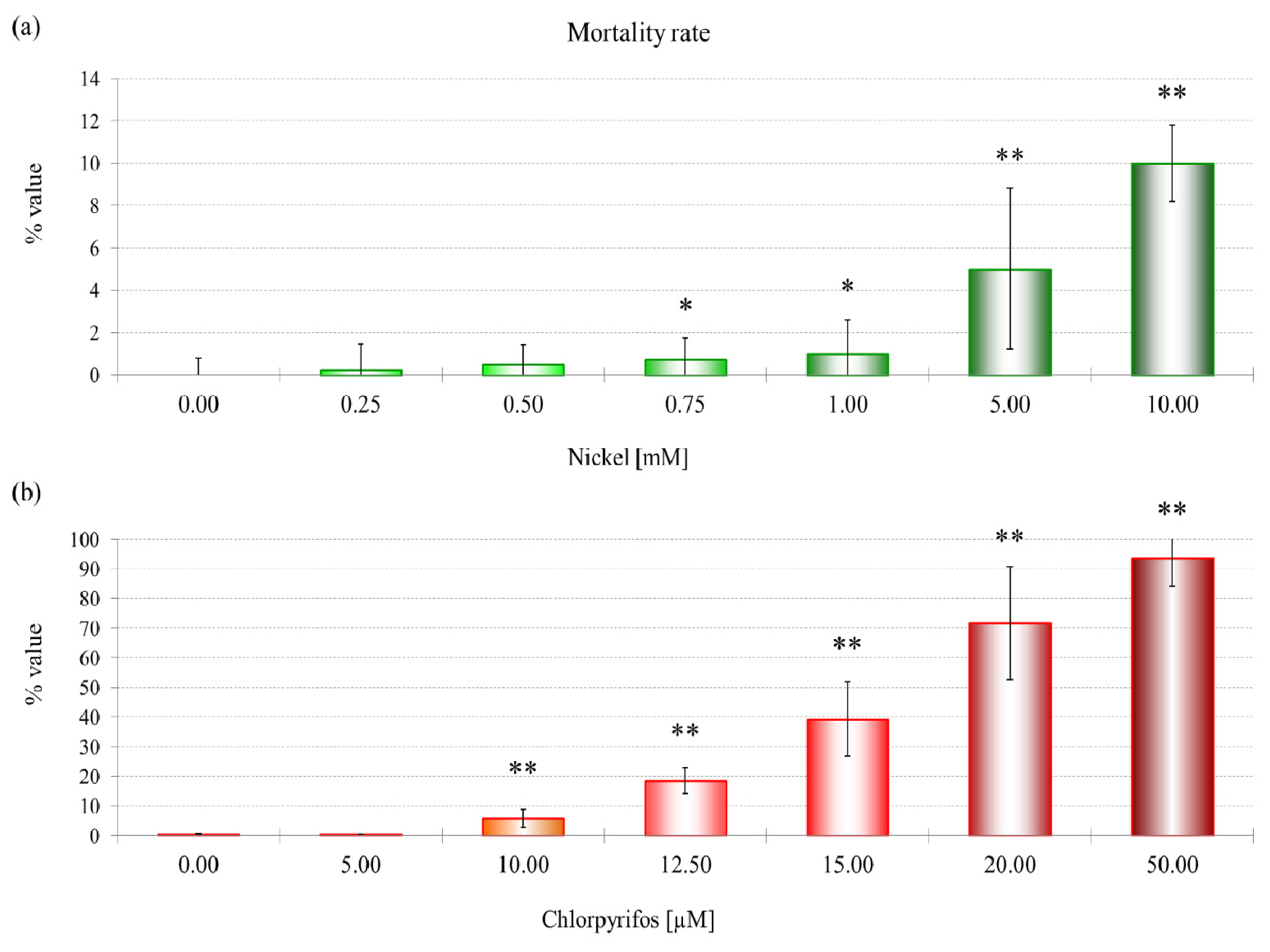
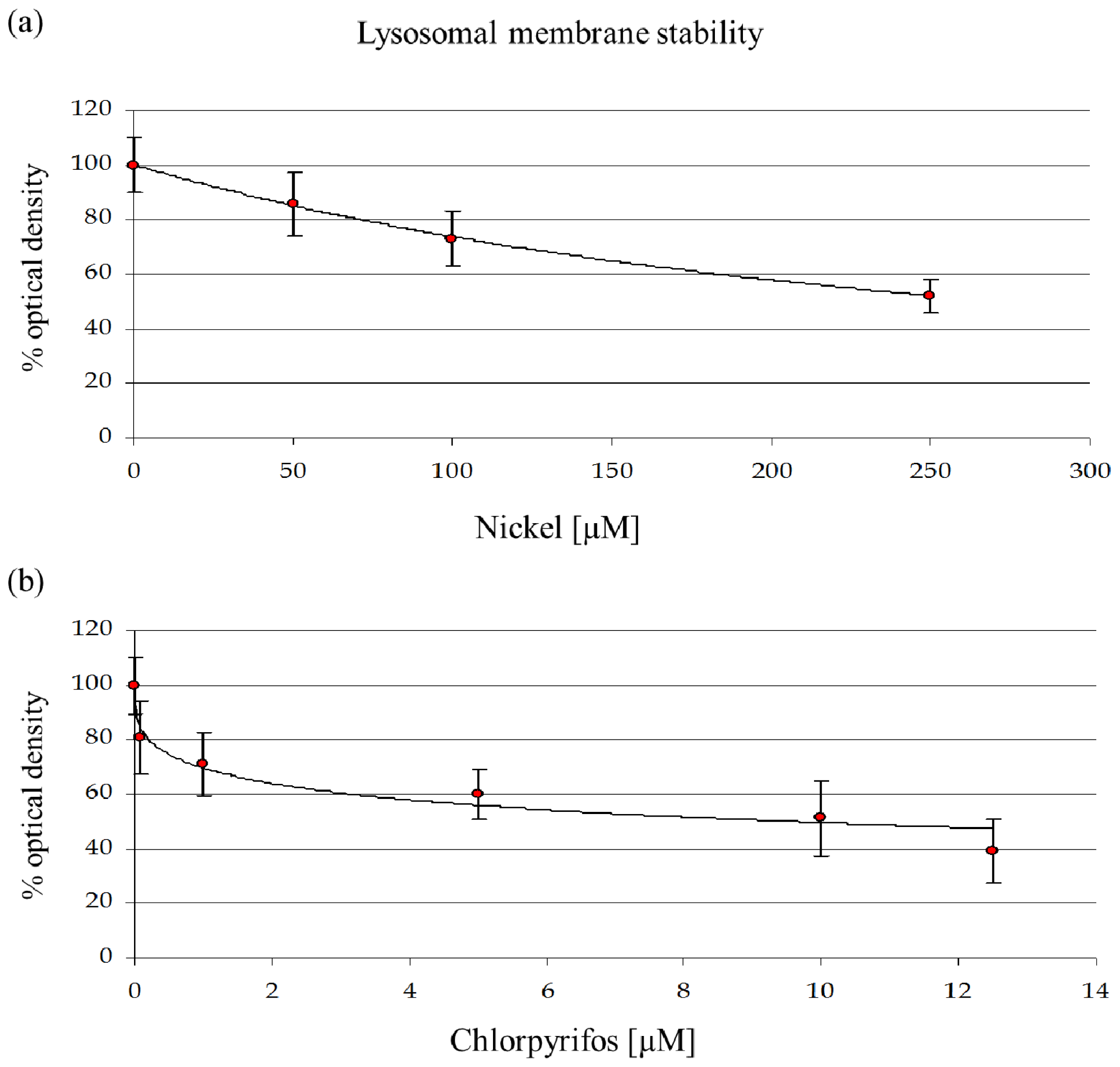
29]. The mortality rate and lysosomal membrane destabilization data (
Figures 1 and
3) revealed that the cytotoxicity increased with an increase in the NiCl
2 concentration. Although the first effects on mortality were observed only after treatment with 0.75 mM NiCl
2, at the subcellular level, LMS and proteomic data show effects at 10% of that concentration.
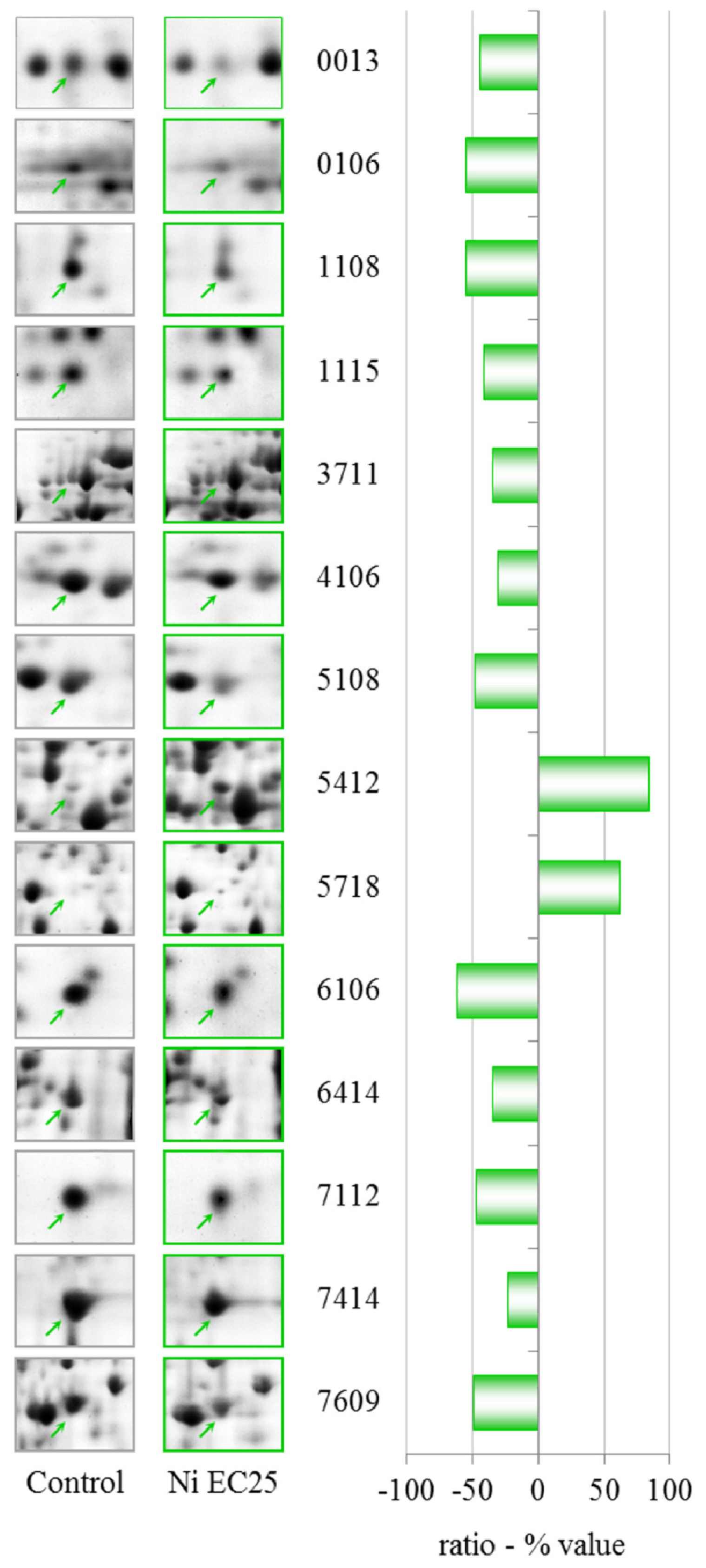
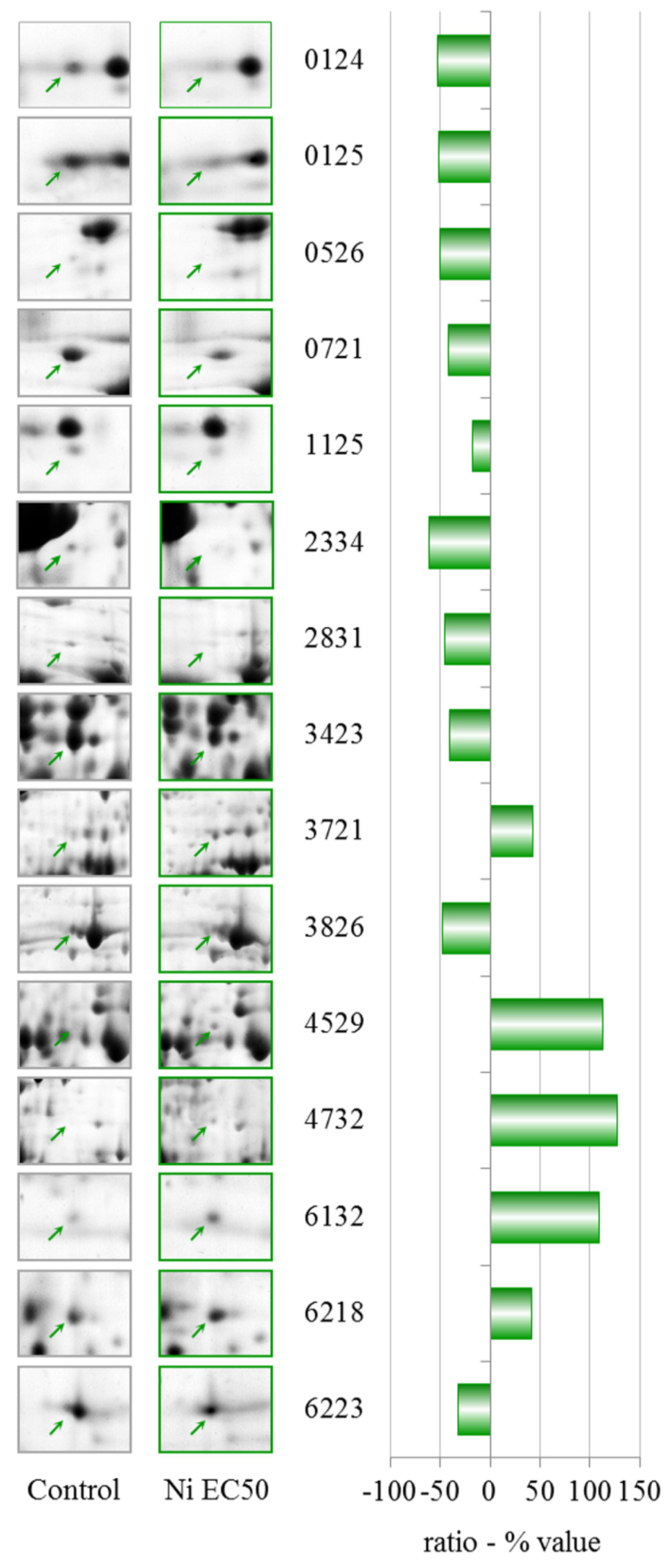
A concentration of 114.6 μM NiCl2, which corresponds to an EC25 for lysosomal membrane destabilization, was chosen for subsequent experiments. The sublethal effects at this concentration were evident, which might indicate that differential protein expression already occurs at this concentration. Moreover, after the sublethal treatments, amoebae could maintain a normal phenotype, which allowed us to extract sufficient amounts of proteins for 2DE experiments. A more toxic concentration of 249.5 μM of NiCl2, corresponding to EC50 for LSM, was chosen for comparison.
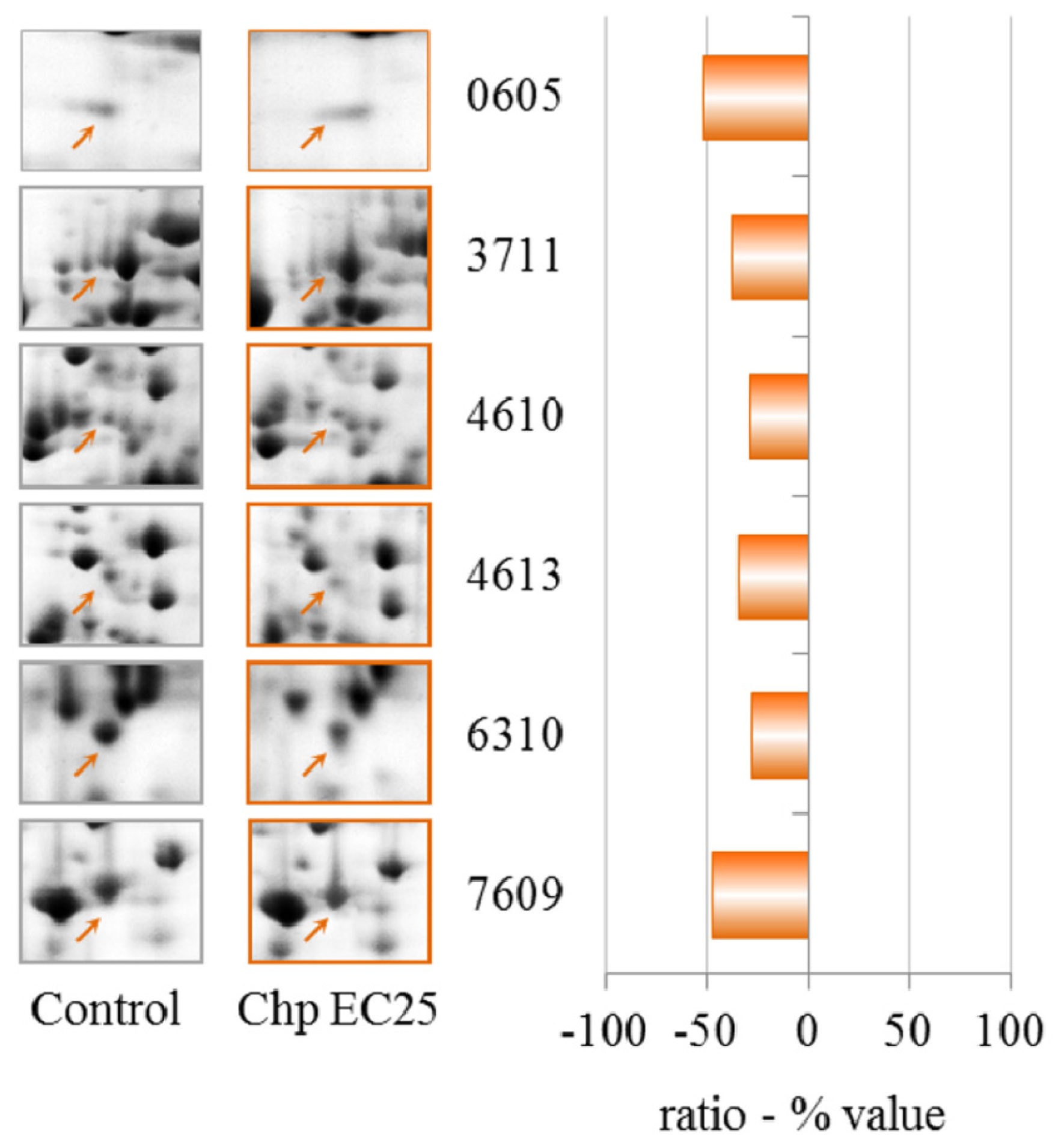
The data in
Table 1 show that clear effects on the proteome can be observed after treatment with NiCl
2 at EC25. Two proteins were up-regulated, succinate dehydrogenase and an isoform of
S-adenosylmethionine synthetase (gi|66803080). The activity of succinate dehydrogenase has been previously observed to be inhibited by cadmium, with the possibility of interfering with energy transport [
30].
S-Adenosylmethionine synthetase (SAM-S) catalyzes the biosynthesis of S-adenosylmethionine from Met and ATP. In plants, the expression of SAM-S is significantly altered in response to salt and drought [
31,
32]. In yeast, the activity of this enzyme is strongly inhibited by heavy metal ions (Cu
2+ and Ag
2+) and ethylenediaminetetraacetic acid (EDTA). Moreover, the expression levels of SAM-S were markedly up-regulated in response to arsenate in rice roots [
33]. Unexpectedly, a different isoform of
S-adenosyl-methionine synthetase (gi|60463691) was down-regulated.
Cyclase-associated proteins (CAPs) are widely distributed and highly conserved proteins that regulate actin remodeling, vesicle trafficking and development in response to cellular signals [
34]. In
D. discoideum, CAPs are essential for the functions of the endo-lysosomal system [
35]. This protein was down-regulated after treatment with CHP at EC25, and both NiCl
2 and CHP EC25 treatments had the clearest effects on endocytosis. The effects on endocytosis are probably linked to the down-regulation of a subunit of the actin-related protein 2/3 (ARP2/3) complex, which plays a major role in actin cytoskeletal regulation.
Glutamate-ammonia ligase was recently identified as a nickel-binding protein by mass spectrometry, and its activity is inhibited by nickel [
36].
Rho GDP dissociation inhibitor (RhoGDI) plays an essential role in the control of a variety of cellular functions through its interactions with Rho family GTPases. RhoGDI is frequently over-expressed in human tumors and chemo-resistant cancer cell lines [
37]. The observed down-regulation due to NiCl
2 could be of great interest and should be studied further.
The down-regulation of LIM-type zinc finger-containing protein after NiCl
2 treatment is not surprising because low concentrations of transition metal have been shown to interfere with DNA transcription and repair. Various effects of heavy metals on zinc finger-containing proteins have been identified, including the displacement of zinc by cadmium or cobalt, the formation of mixed complexes, incomplete coordination of toxic metal ions, and the oxidation of cysteine residues within the metal-binding domain [
38].
The down-regulation of 60S acidic ribosomal protein P2, a ribosomal protein-like and elongation factor 1β, suggests a modulation in protein synthesis caused by the presence of nickel.
After comparing the results of the cytotoxicity tests and protein expression data, it can be seen that when the cells were treated with NiCl
2 concentrations ranging from EC25 to EC50, the cytotoxic effects were linear, but different altered proteins were identified (
Tables 1 and
2). These data indicate that NiCl
2 toxicity cannot be quantitatively assessed by following the expression of single proteins in such a wide concentration range.
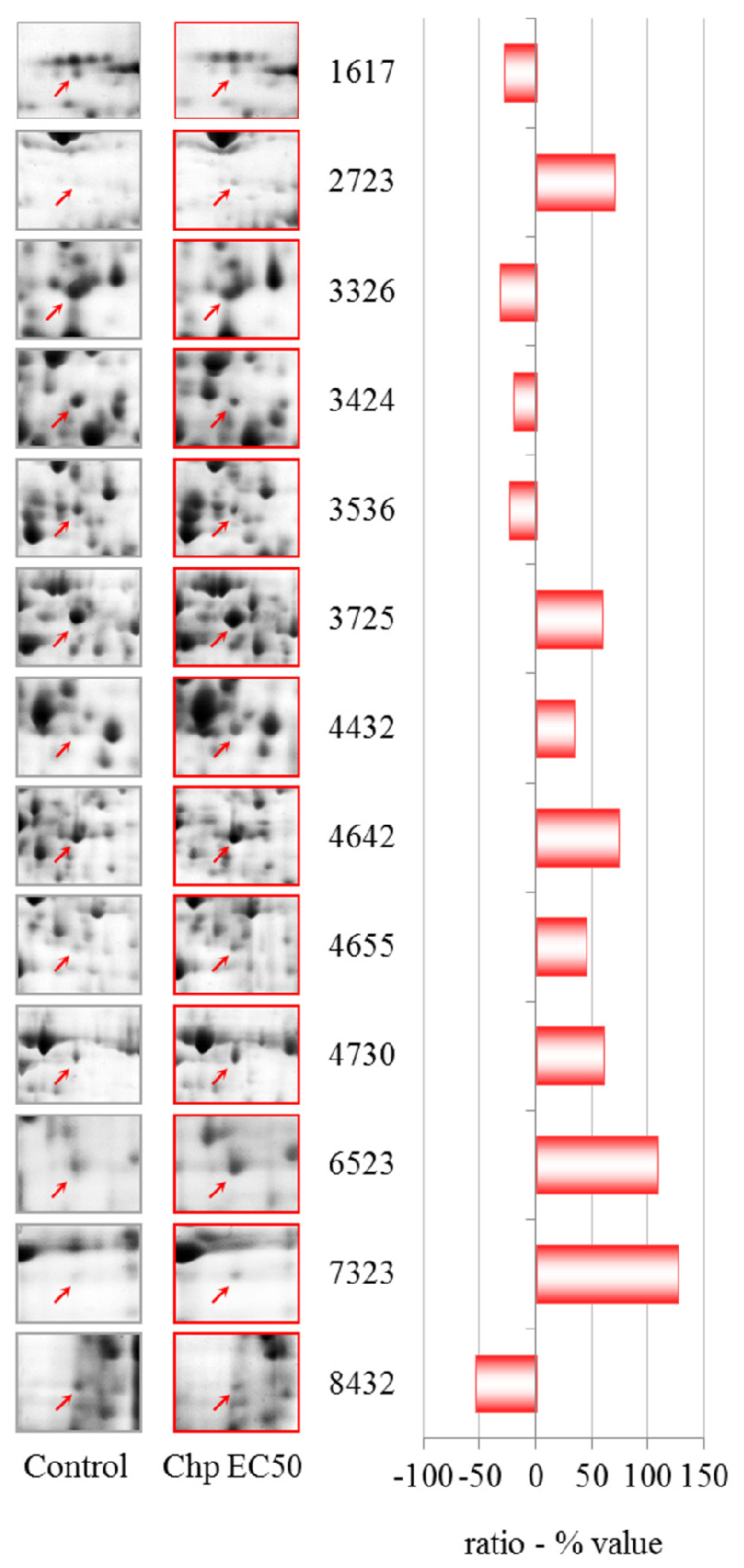
NiCl
2 at EC50 induces different patterns of up-regulation, including an increase in the level of peroxiredoxin. The enzymes in this family consist of sulfhydryl-dependent peroxidases that exhibit high reactivity with hydrogen peroxide and play major roles in peroxide defense, redox cell signaling and metal responses [
39,
40]. Peroxiredoxin has recently been found to have an increased expression in nickel-exposed workers [
41]. Nickel causes increased levels of endogenous cellular hydrogen peroxide and its short-lived reactive oxygen species [
42–
44].
Up-regulation of actin binding protein along with a modulation of the ARP2/3 complex and CAPs substantiates the involvement of the cytoskeleton in the cellular response to nickel. This up-regulation of cytoskeletal proteins in the EC50 treatment could explain the partial recovery of endocytic function. Accordingly, the up-regulation of AAA-ATPase domain-containing protein and a putative epimerase should be investigated further. There is little information about these two proteins, but a member of the AAA-ATPase family has recently been described as a novel anti-apoptotic factor in lung adenocarcinoma cells [
45].
We observed the down-regulation of different proteins after treatment with NiCl
2 at EC50. Major vault protein (MVP), which is the main component of the vault complex, is over-expressed in many multidrug-resistant cancer cell lines, suggesting a possible role for MVP in cell signaling and survival [
46,
47]. Vault complexes are implicated in the cellular response to environmental toxins, but we observed a reduction in the amount of this protein; thus, it is possible that NiCl
2 does not activate this response pathway.
Glucose-regulated protein 94 is a member of the endoplasmic reticulum (ER) chaperone family. In
D. discoideum, it is a constitutively expressed protein that is over-expressed in certain abnormal cellular conditions, including the depletion of glucose and calcium, low oxygen levels and low pH [
48]. We observed down-regulation of this protein after treatment with NiCl
2 at EC50.
We observed the down-regulation of phosphoglycerate kinase after treatment with NiCl2 at EC50. The role of phosphoglycerate kinase in glycolysis is very important for the production of ATP through substrate level phosphorylation. Regulation of this protein is thereby controlled by the energy level of the cell.
Phosphoribosyl pyrophosphate synthetase (PRPS), which was also down-regulated, plays a key role in the biosynthesis of several amino acids and pyrimidines. PRPS is required to convert ribose 5-phosphate into phosphoribosyl pyrophosphate. The latter is a central compound for cellular metabolism and may be considered as a link between carbon and nitrogen metabolism.
NiCl2 induces the down-regulation of the cytosolic glycoprotein FP21 of D. discoideum, which is an orthologue of human and mouse S-phase kinase-associated protein 1. This protein is an essential component of the SCF (SKP1-CUL1-F-box protein) ubiquitin ligase complex, which mediates the ubiquitination of proteins involved in cell cycle progression, signal transduction and transcription.
We observed the down-regulation of the nascent polypeptide-associated complex alpha subunit. This protein prevents short, recently synthesized ribosome-associated polypeptides from inappropriate interactions with cytosolic proteins.
3.2. Chlorpyrifos
The data show a toxic effect of CHP in D. discoideum, an organism that possesses no nervous system. CHP at 9.6 μM already induces the first lethal effects and 50% lysosomal membrane destabilization. Moreover, this compound exerts negative effects on endocytosis.
The proteomic data reveal that CHP at EC25 induces the down-regulation but not up-regulation of proteins. Cyclase-associated proteins are multifunctional proteins that contain several structural domains. Recent studies have suggested important roles for these proteins in linking cell signaling with actin polymerization and highlight their roles in vesicle trafficking and development, as discussed for NiCl2 at EC25.
We observed the down-regulation of the 26S proteasome ATPase 1 subunit. The ubiquitin/26S proteasome pathway is implicated in numerous diseases, including cancer and neurodegenerative diseases [
49]. The 26S proteasome regulatory complex has intrinsic ATPase activity that seems to play an essential role in its function. Presumably, one role of the ATPase is to supply energy continuously for the selective degradation of target proteins by the active 26S proteasome.
In bacteria, yeast and plants, glutamate-ammonia ligase (glutamine synthetase) expression is tightly regulated by the metabolic status of the cell [
50]. A previous study in freshwater crabs showed that glutamine synthetase activity increased after long-term organophosphate exposure [
51]. We observed the down-regulation of this enzyme, indicating that this protein is involved in the response to acute exposure to CHP in
D. discoideum.
Isocitrate dehydrogenase (ICDH) catalyzes the oxidative decarboxylation of isocitrate. Eukaryotes possess two distinct types of ICDH: an NAD-specific enzyme (EC 1.1.1.41), which is present only in the mitochondria, and an NADP-specific enzyme (EC 1.1.1.42), which is found in both the cytoplasm and the mitochondria. The reduced activity of liver mitochondrial ICDH has been correlated with organophosphate toxicity [
52,
53]. There is also evidence correlating the activities of ICDH and the maintenance of the cellular redox state, suggesting that ICDH plays an important role in cellular defenses against oxidative stress [
54].
Aldehyde dehydrogenase catalyzes the oxidation of various aldehydes, using NAD+ as a cofactor [
55]. The enzyme is inhibited by dithiocarbamate fungicides and has previously been described as a suitable detector for these pesticides in the environment [
56,
57]. This enzyme is down-regulated by treatment with CHP at EC25.
Increased transcription of ZPR1-type zinc finger-containing protein has been observed in potato tuber periderm after heat stress [
58]. The deficiency of the zinc finger protein ZPR1 causes neuro-degeneration and defects in transcription in the mouse [
59] and cell cycle progression in HeLa cells [
60]. After treatment with CHP at EC25, we observed the down-regulation of this protein.
The EC50 treatment induces variations in the levels of different protein spots; two of these spots, which were up-regulated, contained two comigrating proteins. In one spot, we identified a tetratricopeptide-like helical domain-containing protein and stress-induced phosphoprotein 1. The other spot contained succinate dehydrogenase and an unknown protein. These spots, which contained comigrating proteins, give no quantitative information because it is impossible to establish which individual protein has altered levels.
The only protein that was down-regulated in both CHP treatments was aldehyde dehydrogenase, which is expected because this protein was previously found to act as a detector of pesticides in the environment [
56,
57].
Among the up-regulated proteins, there is a putative iron regulatory protein (IRP) that acts as a key regulator of cellular iron homoeostasis as a result of the translational control of the expression of a number of iron metabolism-related genes. It is likely that various agents and conditions affect IRP activity, thereby modulating iron and oxygen radical levels in different circumstances [
61].
Previous studies have demonstrated that the up-regulation of transketolase (TKT) might play a critical role in maintaining a reducing environment in the mouse cornea [
62]. Moreover, in the cyanobacterium Anabaena, UV-B upregulated transketolase and protein down-regulation were observed with Cd treatment compared to a control treatment [
63]. The up-regulation that was observed in the present study suggests a role for this protein in the response to CHP.
Another protein that was up-regulated by CHP at EC 50 is annexin VII. Annexins belong to a large family of glycoproteins that bind both Ca
2+ and negatively charged phospholipids and are considered important components of calcium signaling pathways [
64,
65]. Various studies have shown that annexins play important roles in the cell during the response to oxidative stress [
66–
68]. The peroxidase activity that is exhibited by annexins is further exemplified by studies in plants [
69,
70].
β-alanine synthase is an enzyme that catalyzes the final step of pyrimidine catabolism and is required for the biosynthesis of β-alanine in animals. In mammals, β-alanine is a natural antioxidant that is produced by the degradation of uracil or carnosin, which is a polypeptide that is formed by β-alanine and
l-lysine. Moreover, β-alanine is necessary for the production of CoA, which is required for fatty acid metabolism. The treatment of
D. discoideum cells with 9.6 μM CHP induced an increase in ROS production (
Figure S1) with a simultaneous up-regulation of β-alanine synthase, which is the enzyme required for the production of β-alanine. Moreover, as described previously, we found the down-regulation of aldehyde dehydrogenase and
S-adenosylmethionine synthetase (SAMS), both of which are needed in other mechanisms of β-alanine metabolism. We could hypothesize that the attempt by
D. discoideum to modulate a specific pathway for β-alanine production is probably an alternative defense mechanism in response to the oxidative stress that is induced by pesticide treatment.
The expression levels of
S-adenosylmethionine synthetase (SAMS) and a SAM-dependent methyltransferase are markedly up-regulated in response to arsenate in plants [
33]. Conversely, we observed the down-regulation of these proteins after CHP treatment at EC50. This difference requires further investigation.
We observed an increase in phosphopyruvate hydratase (also known as enolase), which is a metalloenzyme that is required for the penultimate step of glycolysis. Increased expression of enolase has recently been reported in stressed plants [
71].
The 60S ribosomal protein L4 is a component of the large ribosomal subunit. Yang
et al. (2005) have show that this protein is required for rRNA processing in mammalian cells, and that decreasing the level of this peptide inhibits the production of several rRNAs [
72].
The exact physiological role of protein phosphatase 2C-like domain containing protein, which we found to be down-regulated after CHP at EC50 treatment, is unclear.
3.3. Mixture
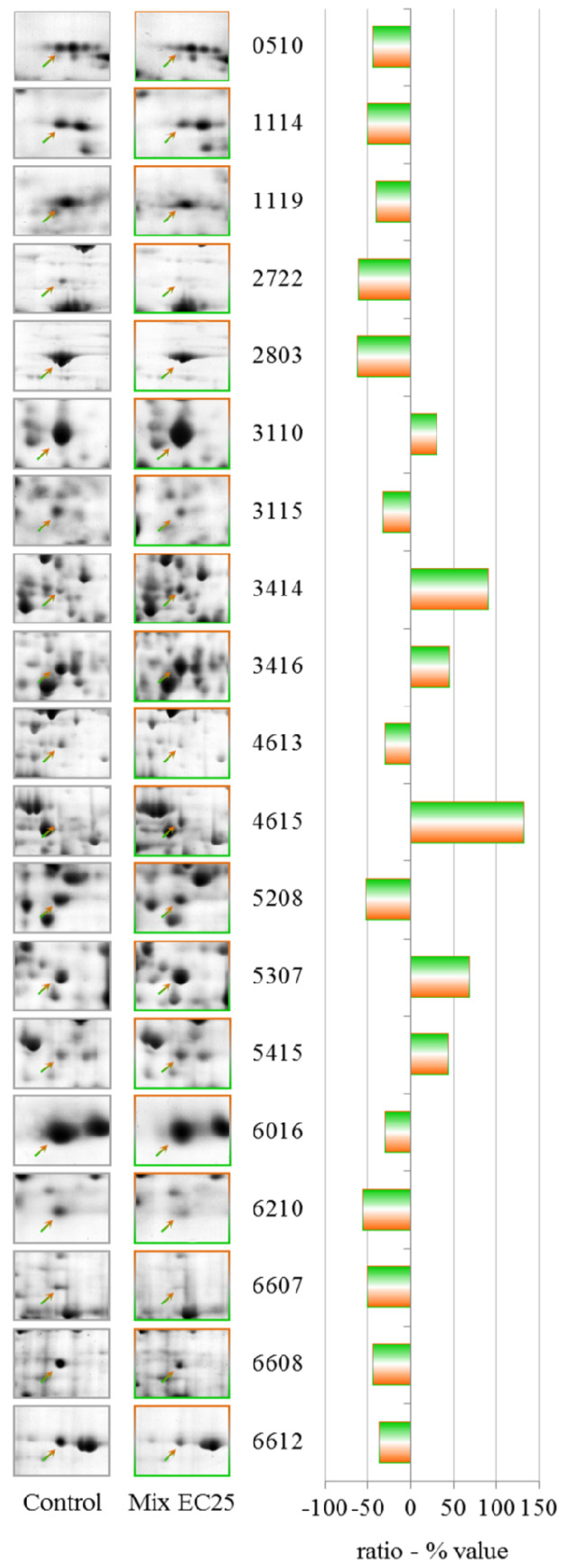
The results have demonstrated that each treatment results in different PESs, and the mixture gives a unique PES, making it difficult to apply PES to the toxicological evaluation of complex environmental matrixes. However, the data show that some proteins that have altered levels after the single toxicant treatments are involved in the cellular response to the mixture. The prominent protein in this class is cyclase-associated protein, which is down-regulated in both single treatments at EC25. Aldehyde dehydrogenase is down-regulated after treatment with CHP at EC25 but up-regulated by treatment with the mixture, along with aldehyde reductase, indicating an attempt by the cell to counteract the formation of lipid peroxide-derived aldehydes that are produced downstream of ROS [
73].
The down-regulation of G β-like protein indicates an involvement of G proteins in the response to the mixture. G proteins represent one of the most prevalent signaling systems in mammalian cells, regulating systems as diverse as sensory perception, cell growth and hormonal regulation.
Phosphoglycerate kinase catalyzes the reversible transfer of a phosphate group from ATP to 3-phosphoglycerate, producing ADP and 1,3-bisphosphoglycerate. This protein is down-regulated by CHP at EC50 but is up-regulated by the mixture.
S-adenosyl-methionine synthetase levels are altered by both NiCl2 EC25 and CHP EC50, and the protein was strongly up-regulated after treatment with the mixture; a putative δ-24-sterol methyltransferase that uses S-adenosyl-methionine as substrate was also up-regulated by the mixture.
Different cytoskeletal proteins, including actin binding protein, myosin II heavy chain, and clathrin heavy chain, were down-regulated by the mixtures, indicating the involvement of the cytoskeleton in the response to toxic substances, despite the fact that treatment with the mixture showed a partial recovery of the endocytic rate.
Isocitrate dehydrogenase (NAD+) levels were down-regulated by the mixture and by CHP at EC25. Aconitase (mitochondrial) has been established as a sensitive target of ROS, and the levels of this enzyme were down-regulated after treatment with the mixture.
ATP citrate synthase catalyzes the first reaction in the Krebs cycle, the conversion of oxaloacetate and acetyl-coenzyme A into citrate and coenzyme A. This reaction is important for energy generation and for carbon assimilation and was down-regulated by the mixture.
We observed the up-regulation of 3-phosphoglycerate dehydrogenase, which catalyzes the transition of 3-phosphoglycerate to 3-phosphohydroxypyruvate. This reaction is the first and rate-limiting step in the phosphorylated pathway of serine biosynthesis; serine is a constituent of proteins and a precursor of several metabolites, including cysteine, glycine and choline.
We observed the down-regulation of the pyruvate dehydrogenase E1 beta subunit. The pyruvate dehydrogenase complex is a nuclear-encoded mitochondrial multi-enzyme complex that catalyzes the overall conversion of pyruvate to acetyl-CoA and CO2 and provides a primary link between glycolysis and the tricarboxylic acid (TCA) cycle.
Another down-regulated protein was glucosamine-6-phosphate isomerase, which catalyzes the conversion of d-glucosamine 6-phosphate (GlcN6P) to d-fructose 6-phosphate and ammonia. This reaction is the last step in the pathway for N-acetylglucosamine utilization in bacteria or fungi.
Another interesting protein that was strongly up-regulated by the mixture was a putative transport protein that should be studied further to clarify its involvement in the response to the toxic mixture.
Based on these results, it is possible to investigate the molecular mechanisms of cytotoxicity on a proteomic scale by using a proteomic approach, despite the fact that each treatment has a different PES. In particular, this approach, beyond the simultaneous quantitative observation of proteins that are known to be stress responsive in a single experiment, allows us to identify novel proteins that have never previously been observed to change in level after toxic treatments.