Differential Expression Profile of MicroRNAs during Differentiation of Cardiomyocytes Exposed to Polychlorinated Biphenyls
Abstract
:1. Introduction
2. Results and Discussion
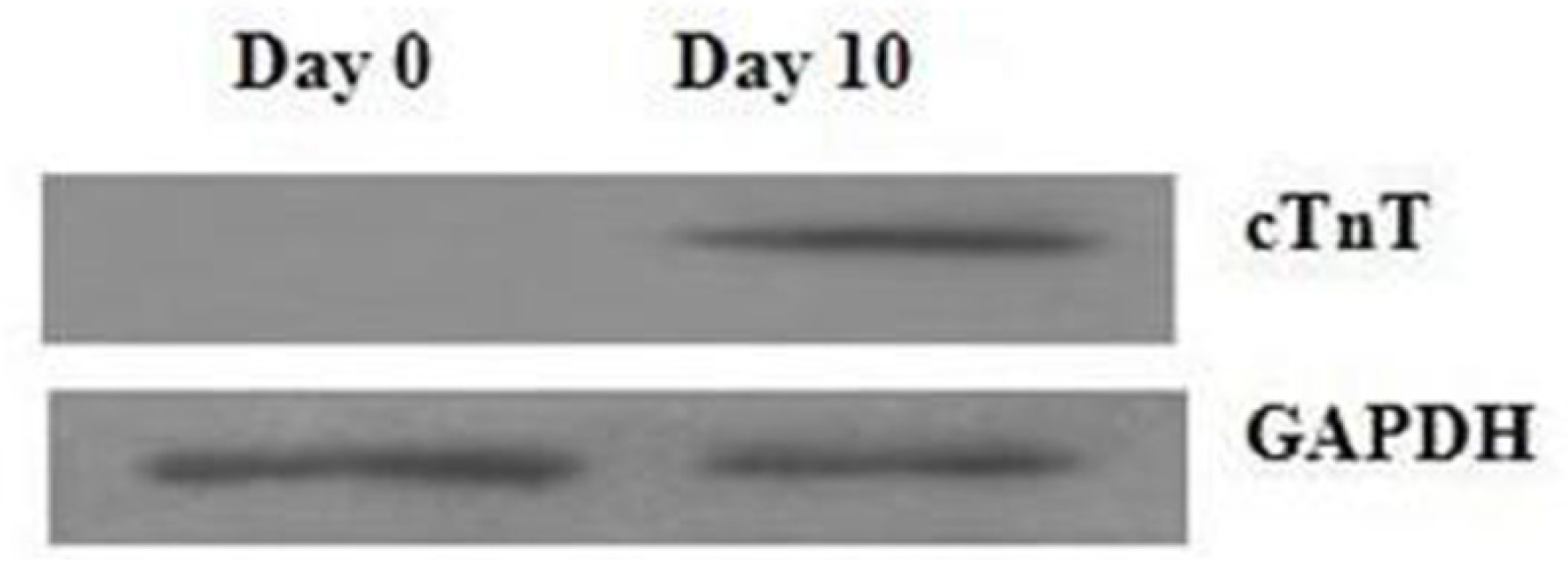
2.1. Assessment of Cell Differentiation
2.2. Morphology of P19 Cells during Differentiation
2.3. miRNA Expression Profiles in P19 Cells Exposed to PCBs Relative to Control Cells
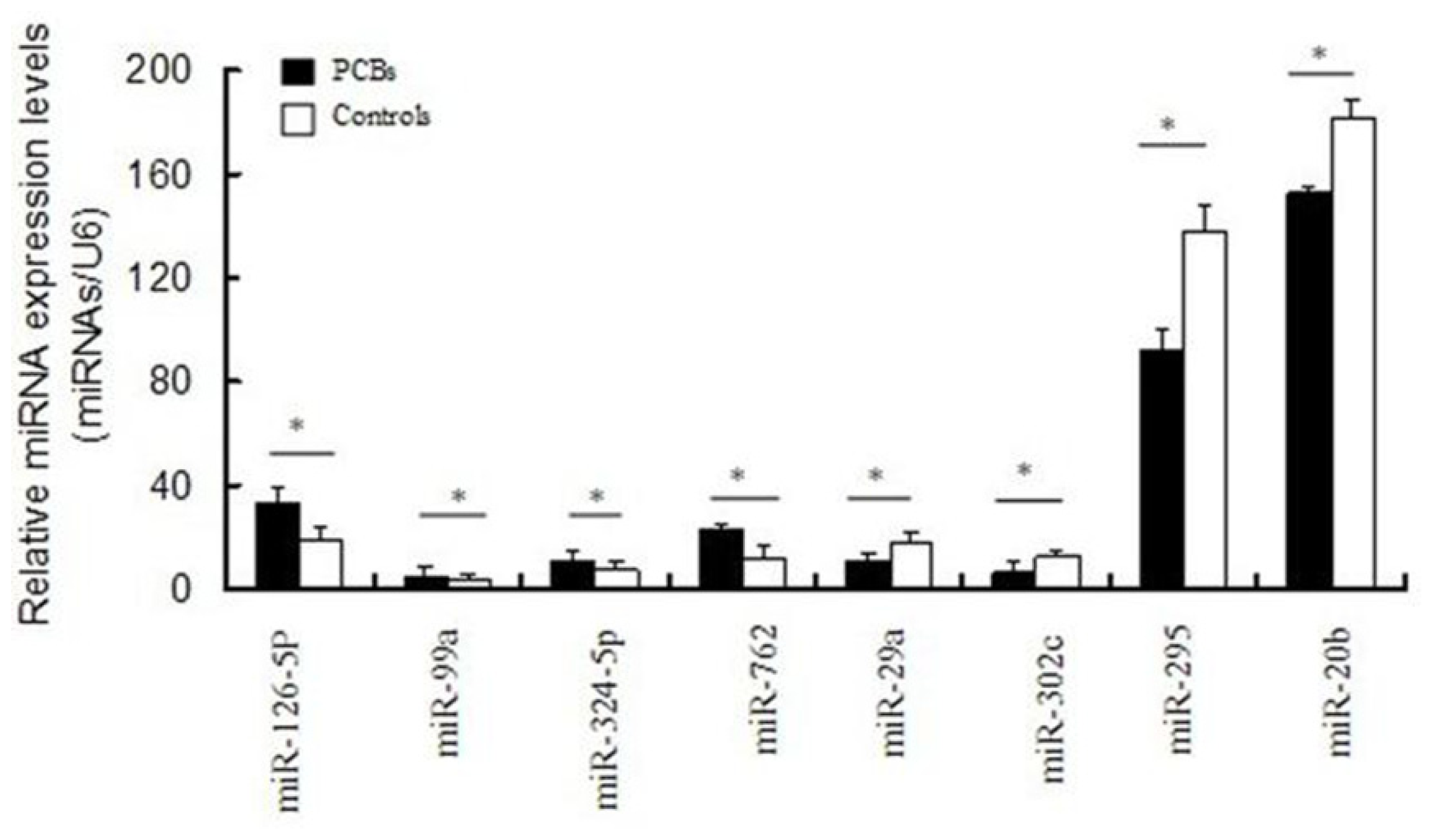
2.4. Validation of Differentially Expressed miRNAs
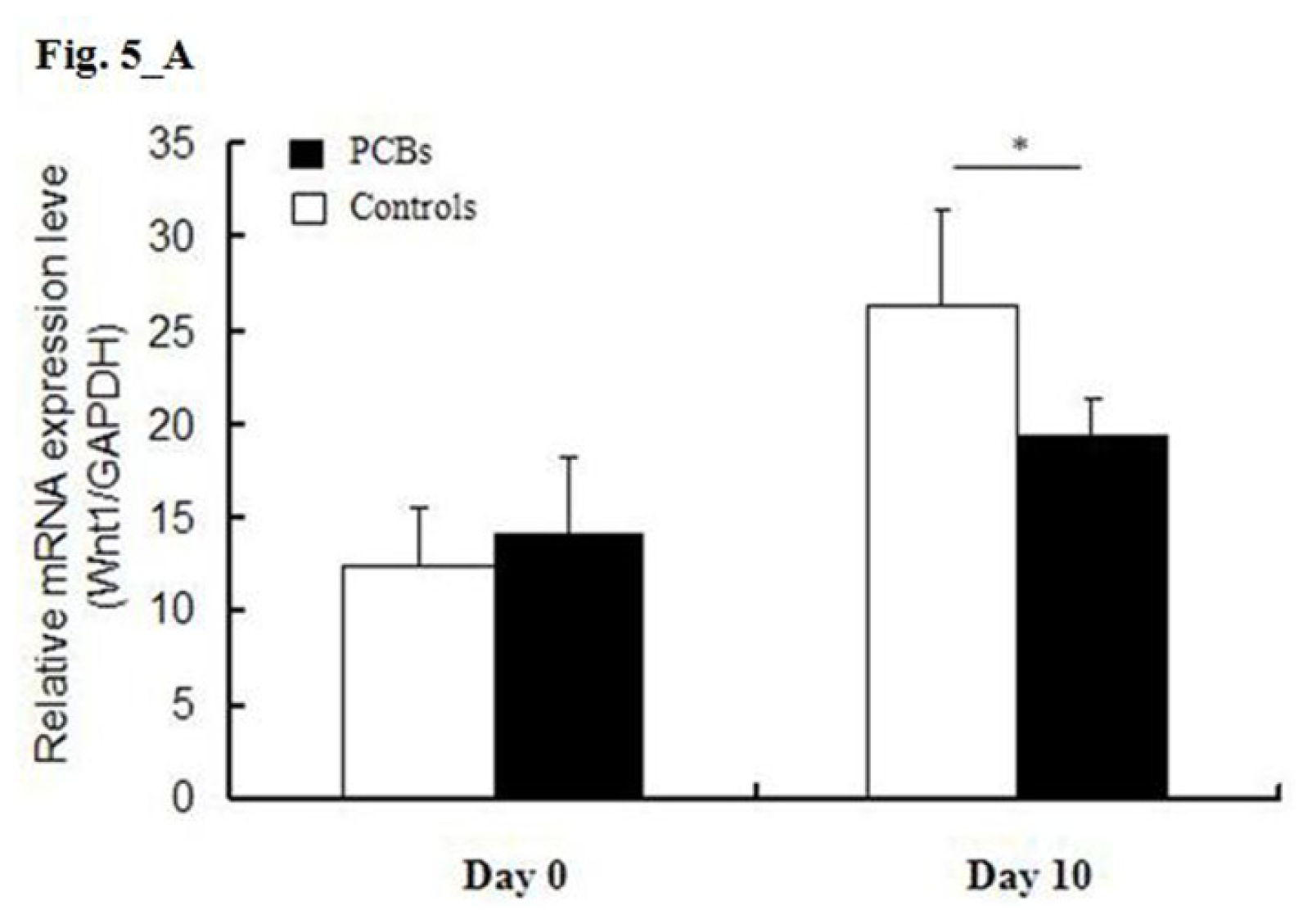
2.5. miRNA Target Predictions
2.6. Discussion
3. Experimental Methods
3.1. Cell Culture
3.2. Western Blotting
3.3. Total RNA Preparation
3.4. MiRNA Microarray
3.5. Validation of Microarray Experiments
3.6. MiRNA Target Predictions and Real-Time RT-PCR
4. Conclusion
Acknowledgements
References
- Crinnion, W.J. Polychlorinated biphenyls: Persistent pollutants with immunological, neurological, and endocrinological consequences. Altern. Med. Rev 2011, 16, 5–13. [Google Scholar]
- Han, S.G.; Eum, S.Y.; Toborek, M.; Smart, E.; Hennig, B. Polychlorinated biphenyl-induced VCAM-1 expression is attenuated in aortic endothelial cells isolated from caveolin-1 deficient mice. Toxicol. Appl. Pharmacol 2010, 246, 74–82. [Google Scholar]
- Hong, Y.; Chunhong, Z.; Xiaoxiong, Z. Investigation of pollution characteristics of polychlorinated biphenyls in the typical drinking water sources in Jiangsu Province, China. Environ. Monit. Assess 2009, 158, 573–579. [Google Scholar]
- Kopf, P.G.; Walker, M.K. Overview of developmental heart defects by dioxins, PCBs, and pesticides. J. Environ. Sci. Health C 2009, 27, 276–285. [Google Scholar]
- Goncharov, A.; Bloom, M.; Pavuk, M.; Birman, I.; Carpenter, D.O. Blood pressure and hypertension in relation to levels of serum polychlorinated biphenyls in residents of Anniston, Alabama. J. Hypertens 2010, 28, 2053–2060. [Google Scholar]
- Van Rooij, E.; Olson, E.N. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Invest 2007, 117, 2369–2376. [Google Scholar]
- Reymann, S.; Borlak, J. Transcriptome profiling of human hepatocytes treated with Aroclor 1254 reveals transcription factor regulatory networks and clusters of regulated genes. BMC Genomics 2006, 7, 217. [Google Scholar]
- Van der Heyden, M.A.; Defize, L.H. Twenty one years of P19 cells: What an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc. Res 2003, 58, 292–302. [Google Scholar]
- Callis, T.E.; Pandya, K.; Seok, H.Y.; Tang, R.H.; Tatsuguchi, M.; Huang, Z.P.; Chen, J.F.; Deng, Z.; Gunn, B.; Shumate, J.; et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest 2009, 119, 2772–2786. [Google Scholar]
- Cheng, Y.; Liu, X.; Zhang, S.; Lin, Y.; Yang, J.; Zhang, C. MicroRNA-21 protects against the H2O2-induced injury on cardiac myocytes via its target gene PDCD4. J. Mol. Cell. Cardiol 2009, 47, 5–14. [Google Scholar]
- Dong, S.; Cheng, Y.; Yang, J.; Li, J.; Liu, X.; Wang, X.; Wang, D.; Krall, T.J.; Delphin, E.S.; Zhang, C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem 2009, 284, 29514–29525. [Google Scholar]
- Schwartz, R.J.; Olson, E.N. Building the heart piece by piece: Modularity of cis-elements regulating Nkx2-5 transcription. Development 1999, 126, 4187–4192. [Google Scholar]
- Ma, Q.; Zhou, B.; Pu, W.T. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev. Biol. 2008, 323, 98–104. [Google Scholar]
- Cohen, E.D.; Tian, Y.; Morrisey, E.E. Wnt signaling: An essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development 2008, 135, 789–798. [Google Scholar]
- Kwon, C.; Arnold, J.; Hsiao, E.C.; Taketo, M.M.; Conklin, B.R.; Srivastava, D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc. Natl. Acad. Sci. USA 2007, 104, 10894–10899. [Google Scholar]
- Maye, P.; Zheng, J.; Li, L.; Wu, D. Multiple mechanisms for Wnt11-mediated repression of the canonical Wnt signaling pathway. J. Biol. Chem 2004, 279, 24659–24665. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar]
- Yamasaki, F.; Zhang, D.; Bartholomeusz, C.; Sudo, T.; Hortobagyi, G.N.; Kurisu, K.; Ueno, N.T. Sensitivity of breast cancer cells to erlotinib depends on cyclin-dependent kinase 2 activity. Mol. Cancer Ther 2007, 6, 2168–2177. [Google Scholar]
- miRBase Sequence database. Available online: http://microrna.sanger.ac.uk/sequences/index.shtml accessed on 9 March 2012.
- TargetScan and PicTar target prediction databases. Available online: http://pictar.mdc-berlin.de/cgi-bin/new_PicTar_mouse.cgi accessed on 9 March 2012.
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC T Method. Methods 2001, 25, 402–408. [Google Scholar]


| miRNA | Regulation | Log2 | p-Value |
|---|---|---|---|
| mmu-miR-99a | Up | 1.74 | 0.0060 |
| mmu-miR-214* | Up | 1.51 | 0.0365 |
| mmu-miR-345-5p | Up | 1.57 | 0.0128 |
| mmu-miR-126-5p | Up | 2.89 | 0.0338 |
| mmu-miR-762 | Up | 2.20 | 0.0242 |
| mmu-miR-324-5p | Up | 1.90 | 0.0071 |
| mmu-miR-181a | Up | 1.76 | 0.0318 |
| mmu-miR-302b* | Up | 2.83 | 0.0404 |
| mmu-miR-155 | Up | 1.97 | 0.0333 |
| mmu-miR-33 | Up | 1.90 | 0.0487 |
| mmu-miR-208a | Up | 2.13 | 0.0465 |
| mmu-miR-10a | Up | 1.84 | 0.0226 |
| mmu-miR-154 | Up | 1.64 | 0.0392 |
| mmu-miR-500 | Up | 1.53 | 0.0079 |
| mmu-miR-29a | Down | 0.60 | 0.0139 |
| mmu-miR-302c | Down | 0.65 | 0.0093 |
| mmu-miR-302b | Down | 0.73 | 0.0249 |
| mmu-miR-293 | Down | 0.45 | 0.0349 |
| mmu-miR-294 | Down | 0.52 | 0.0284 |
| mmu-miR-295 | Down | 0.56 | 0.0410 |
| mmu-miR-7a | Down | 0.62 | 0.0435 |
| mmu-miR-302a* | Down | 0.67 | 0.0203 |
| mmu-miR-1949 | Down | 0.58 | 0.0337 |
| mmu-miR-21 | Down | 0.63 | 0.0039 |
| mmu-miR-494 | Down | 0.54 | 0.0165 |
| mmu-miR-20b | Down | 0.55 | 0.0349 |
| mmu-miR-200b | Down | 0.14 | 0.0021 |
| mmu-miR-136 | Down | 0.37 | 0.0346 |
| mmu-miR-141 | Down | 0.05 | 0.0249 |
| mmu-miR-194 | Down | 0.02 | 0.0147 |
| mmu-miR-29b | Down | 0.17 | 0.0350 |
| mmu-miR-32 | Down | 0.48 | 0.0075 |
| mmu-miR-710 | Down | 0.29 | 0.0120 |
| mmu-miR-290-3p | Down | 0.35 | 0.0204 |
| mmu-miR-2133 | Down | 0.45 | 0.0072 |
| mmu-miR-18b | Down | 0.61 | 0.0213 |
| mmu-miR-101a | Down | 0.65 | 0.0273 |
| mmu-miR-326 | Down | 0.47 | 0.0095 |
| mmu-miR-669c | Down | 0.35 | 0.0428 |
| mmu-miR-302d | Down | 0.58 | 0.0113 |
| mmu-miR-2141 | Down | 0.25 | 0.0286 |
| mmu-miR-183 | Down | 0.60 | 0.0151 |
| mmu-miR-363 | Down | 0.56 | 0.0174 |
| mmu-miR-706 | Down | 0.64 | 0.0216 |
| mmu-miR-290-3p | Down | 0.34 | 0.0439 |
| Gene name | RT primer |
|---|---|
| mmu-miR-126-5p | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACCGCGTA 3′ | |
| mmu-miR-99a | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACCACAAG 3′ | |
| mmu-miR-324-5p | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACACACCA 3′ | |
| mmu-miR-762 | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACGCTCTG 3′ | |
| mmu-miR-29a | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACTAACCG 3′ | |
| mmu-miR-302c | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACCCACTG 3′ | |
| mmu-miR-295 | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACAGACTCAA 3′ | |
| mmu-miR-20b | 5′ GTCGTATCCAGTGCGTGTCGTGGAGTC |
| GGCAATTGCACTGGATACGACCTACCT 3′ | |
| U6 | 5′ CGCTTCACGAATTTGCGTGTCAT 3′ |
| Gene name | Primers | Product size (bp) |
|---|---|---|
| mmu-miR-126-5p | F: 5′ GGGGCATTATTACTTTTGG 3′ | 65 |
| R: 5′ CAGTGCGTGTCGTGGA 3′ | ||
| mmu-miR-99a | F: 5′ GATAACCCGTAGATCCGAT 3′ | 65 |
| R: 5′ CAGTGCGTGTCGTGGA 3′ | ||
| mmu-miR-324-5p | F: 5′ CGCATCCCCTAGGGCA 3′ | 61 |
| R: 5′ GTGCGTGTCGTGGAGTCG 3′ | ||
| mmu-miR-762 | F: 5′ ATGCTGGGGCCGGGA 3′ | 59 |
| R: 5′ GTGCGTGTCGTGGAGTCG 3′ | ||
| mmu-miR-29a | F: 5′ GGGTAGCACCATCTGAAAT 3′ | 62 |
| R: 5′ TGCGTGTCGTGGAGTC 3′ | ||
| mmu-miR-302c | F: 5′ GGCAAGTGCTTCCATGTTT 3′ | 65 |
| R: 5′ CAGTGCGTGTCGTGGAGT 3′ | ||
| mmu-miR-295 | F: 5′ GGGGGAAAGTGCTACTACTT 3′ | 68 |
| R: 5′ CAGTGCGTGTCGTGGAG 3′ | ||
| mmu-miR-20b | F: 5′ GGACAAAGTGCTCATAGTGC 3′ | 66 |
| R: 5′ CAGTGCGTGTCGTGGAGT 3′ | ||
| U6 | F: 5′ GCTTCGGCAGCACATATACTAAAAT 3′ | 89 |
| R: 5′ CGCTTCACGAATTTGCGTGTCAT 3′ |
| Gene name | Primers | Product size (bp) |
|---|---|---|
| Wnt1 | F: 5′ GGTTTCTACTACGTTGCTACTGG 3′ | 121 |
| R: 5′ GGAATCCGTCAACAGGTTCGT 3′ | ||
| GSK3β | F: 5′ TGGCAGCAAGGTAACCACAG 3′ | 189 |
| R: 5′ CGGTTCTTAAATCGCTTGTCCTG 3′ | ||
| NKX2.5 | F: 5′ CCTGCGGCCTCTACATGA 3′ | 222 |
| R: 5′ AGGGTCTCACCAGCAGGA 3′ | ||
| GAPDH | F: 5′ TTCACCACCATGGAGAAGGC 3′ | 237 |
| R: 5′ GGCATGGACTGTGGTCATGA 3′ |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhu, C.; Yu, Z.-B.; Zhu, J.-G.; Hu, X.-S.; Chen, Y.-L.; Qiu, Y.-F.; Xu, Z.-F.; Qian, L.-M.; Han, S.-P. Differential Expression Profile of MicroRNAs during Differentiation of Cardiomyocytes Exposed to Polychlorinated Biphenyls. Int. J. Mol. Sci. 2012, 13, 15955-15966. https://doi.org/10.3390/ijms131215955
Zhu C, Yu Z-B, Zhu J-G, Hu X-S, Chen Y-L, Qiu Y-F, Xu Z-F, Qian L-M, Han S-P. Differential Expression Profile of MicroRNAs during Differentiation of Cardiomyocytes Exposed to Polychlorinated Biphenyls. International Journal of Molecular Sciences. 2012; 13(12):15955-15966. https://doi.org/10.3390/ijms131215955
Chicago/Turabian StyleZhu, Chun, Zhang-Bin Yu, Jin-Gai Zhu, Xiao-Shan Hu, Yu-Lin Chen, Yu-Fang Qiu, Zheng-Feng Xu, Lin-Mei Qian, and Shu-Ping Han. 2012. "Differential Expression Profile of MicroRNAs during Differentiation of Cardiomyocytes Exposed to Polychlorinated Biphenyls" International Journal of Molecular Sciences 13, no. 12: 15955-15966. https://doi.org/10.3390/ijms131215955




