Carbohydrate Stress Affecting Fruitlet Abscission and Expression of Genes Related to Auxin Signal Transduction Pathway in Litchi
Abstract
:1. Introduction
2. Results
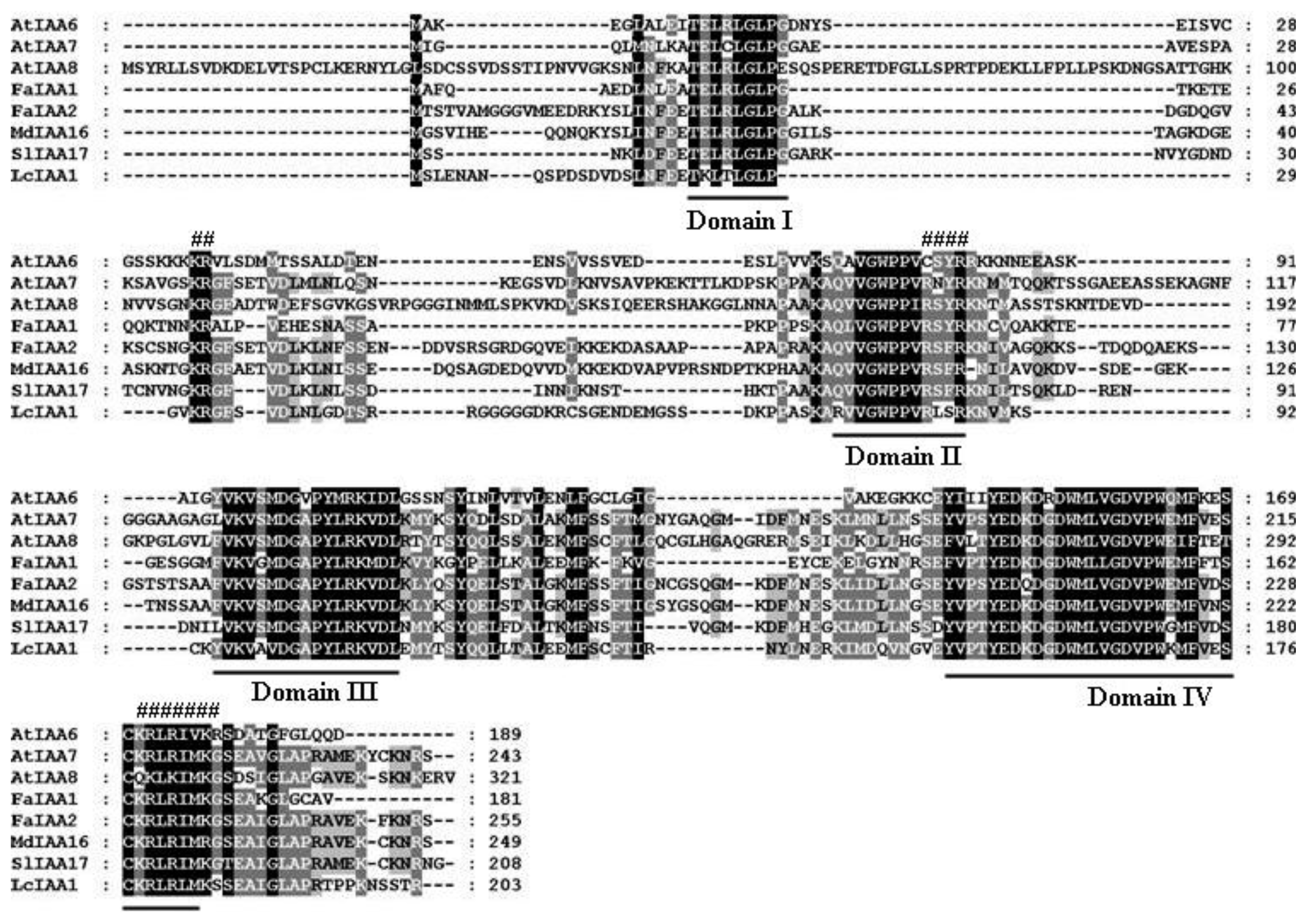
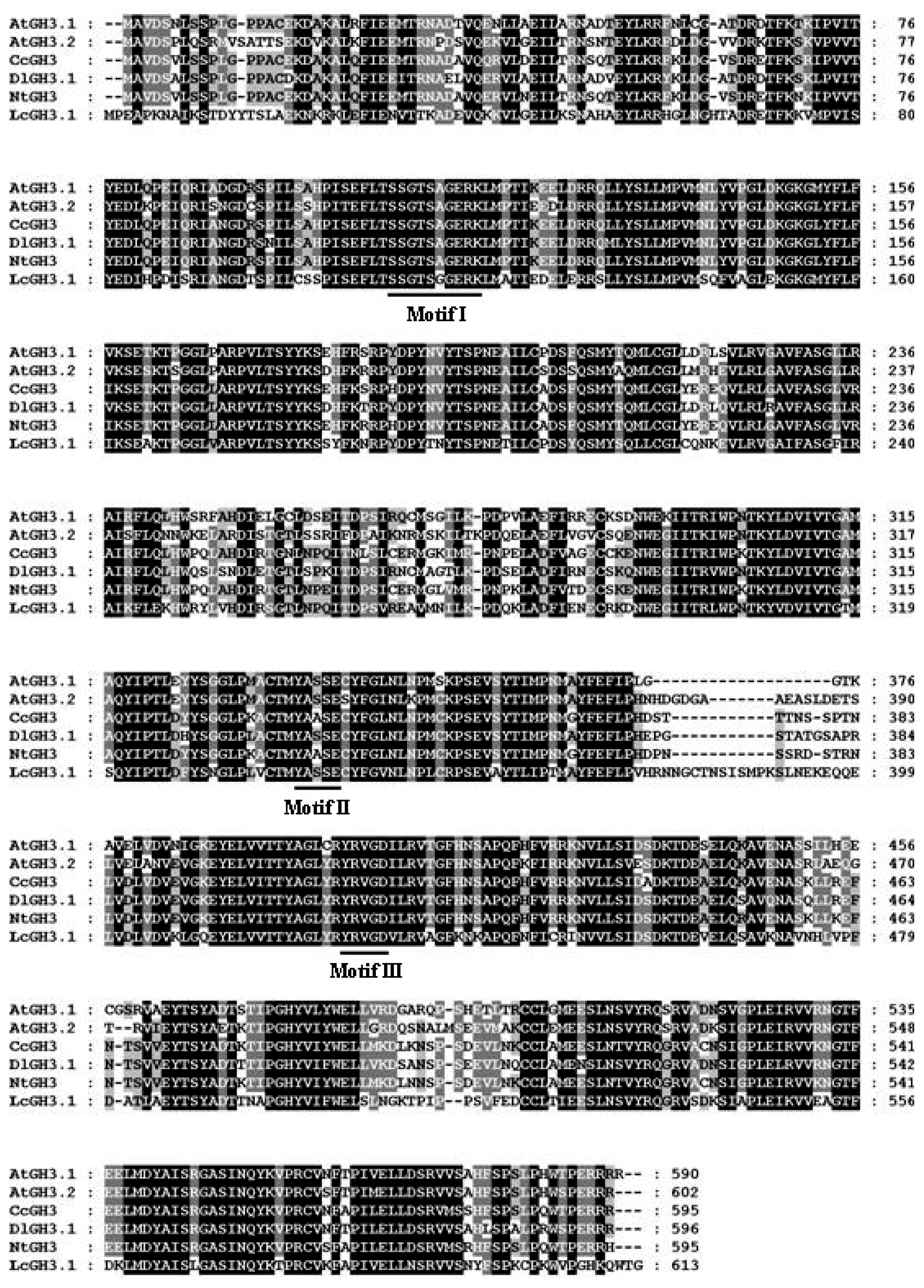
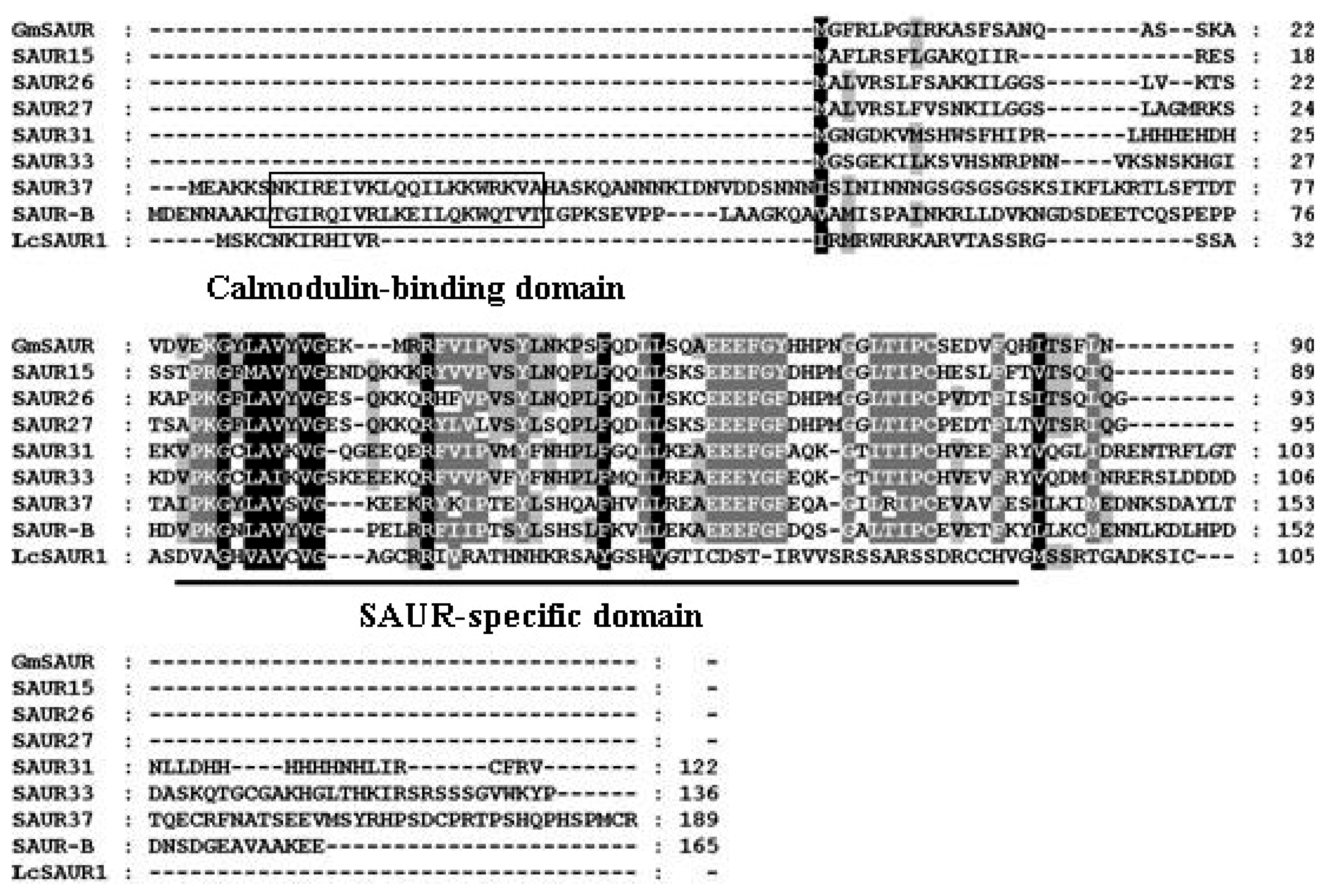
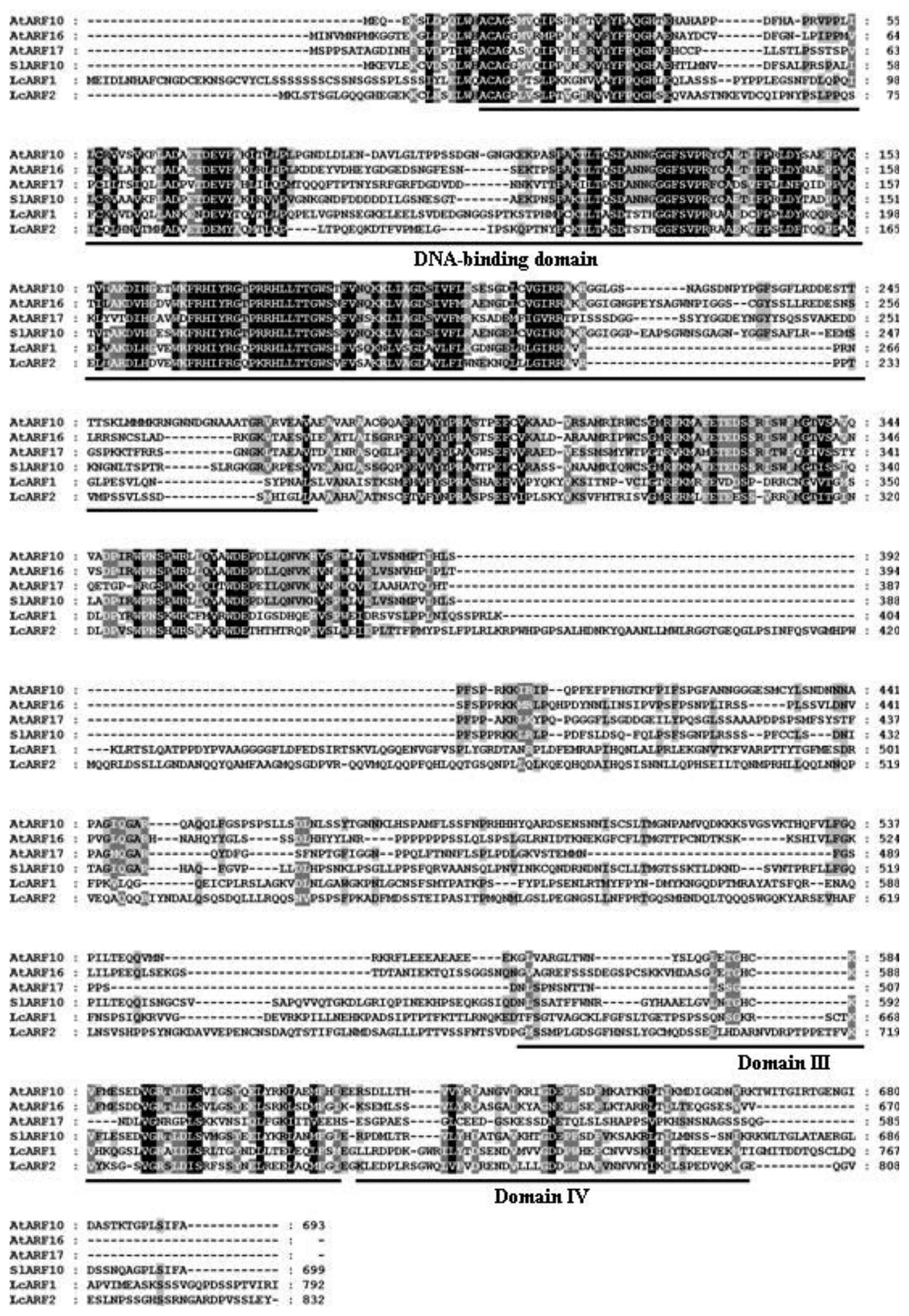
2.1. Isolation of AUX/IAA, GH3, SAUR and ARF Genes from Litchi Fruit
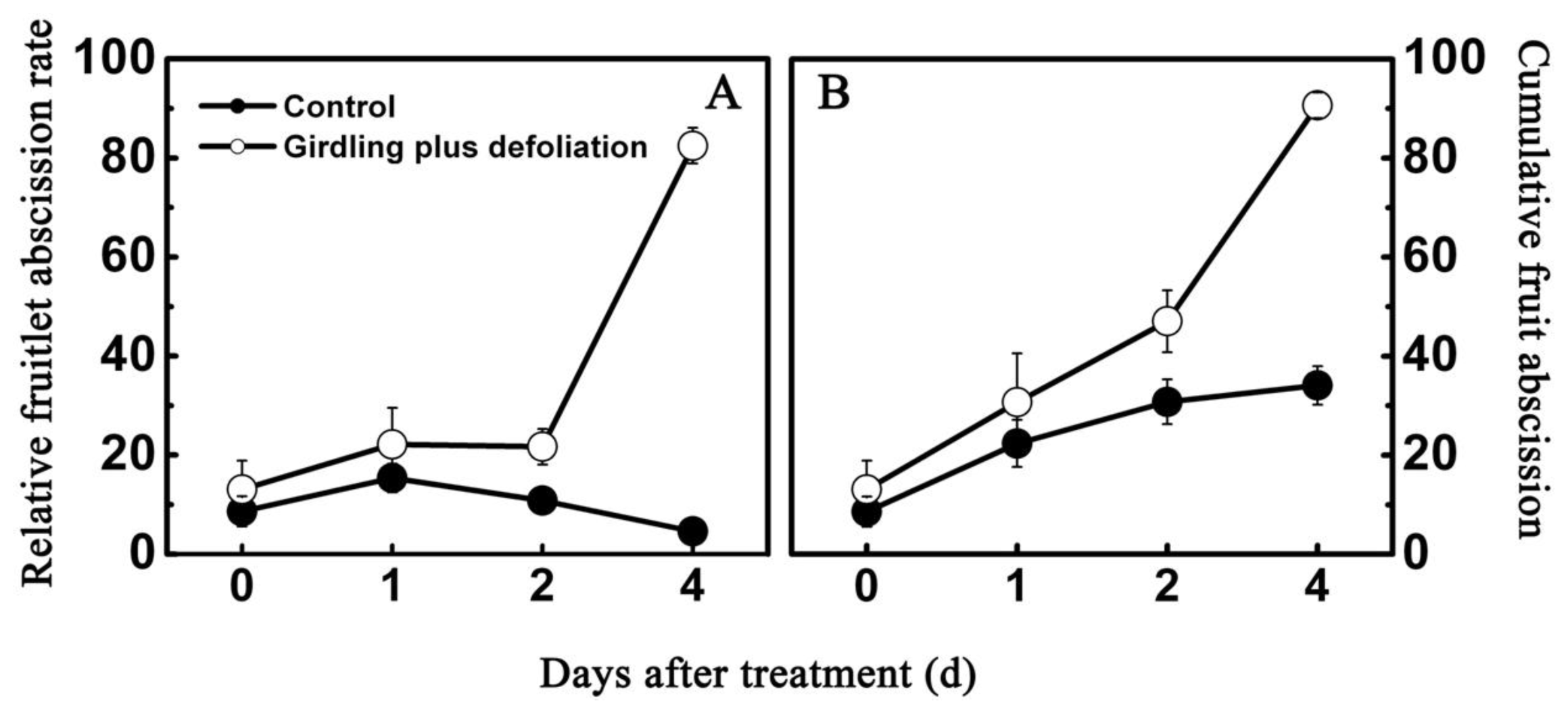
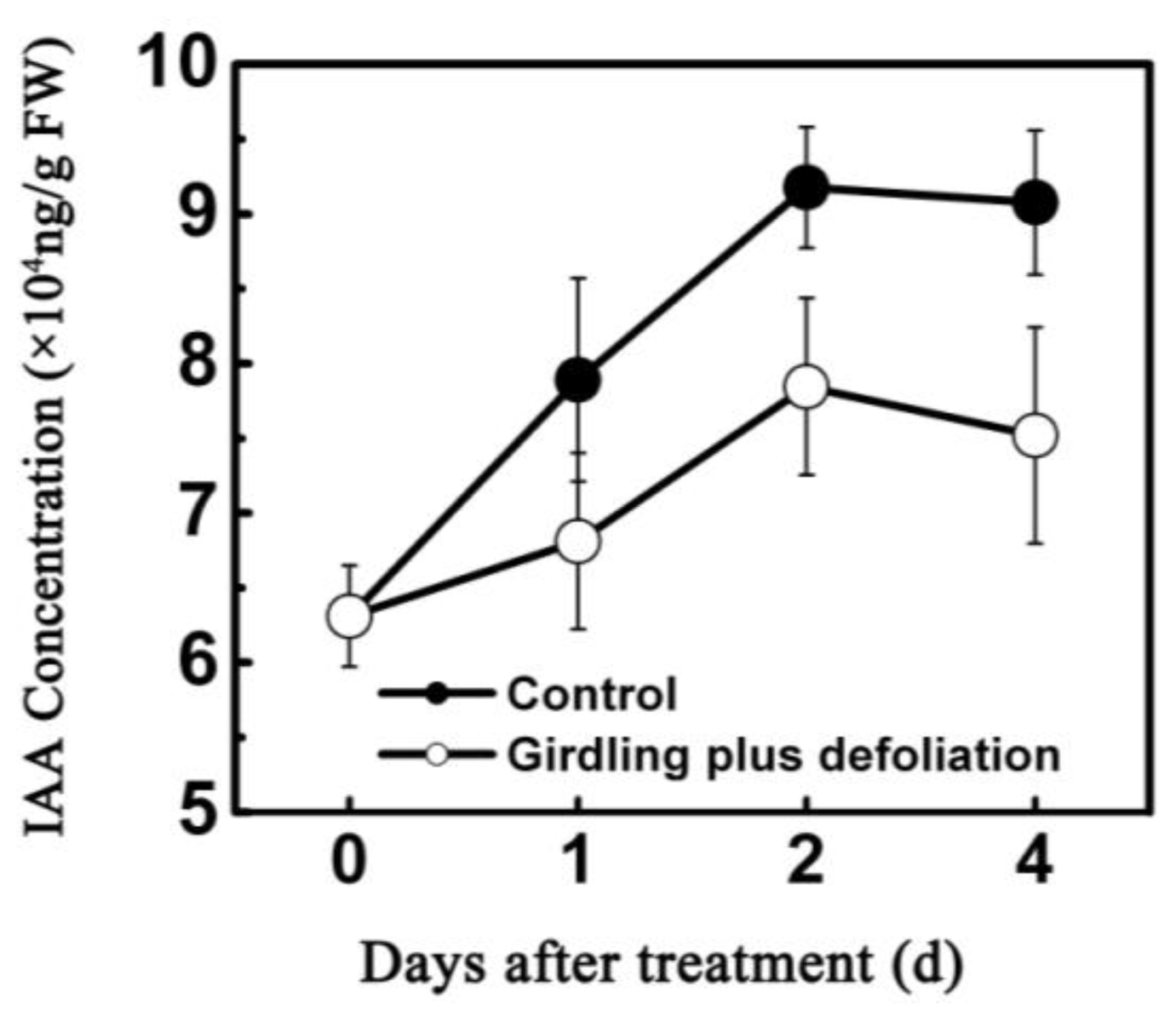
2.2. Effect of Girdling Plus Defoliation on the Abscission of Litchi Fruitlet and Endogenous IAA Content
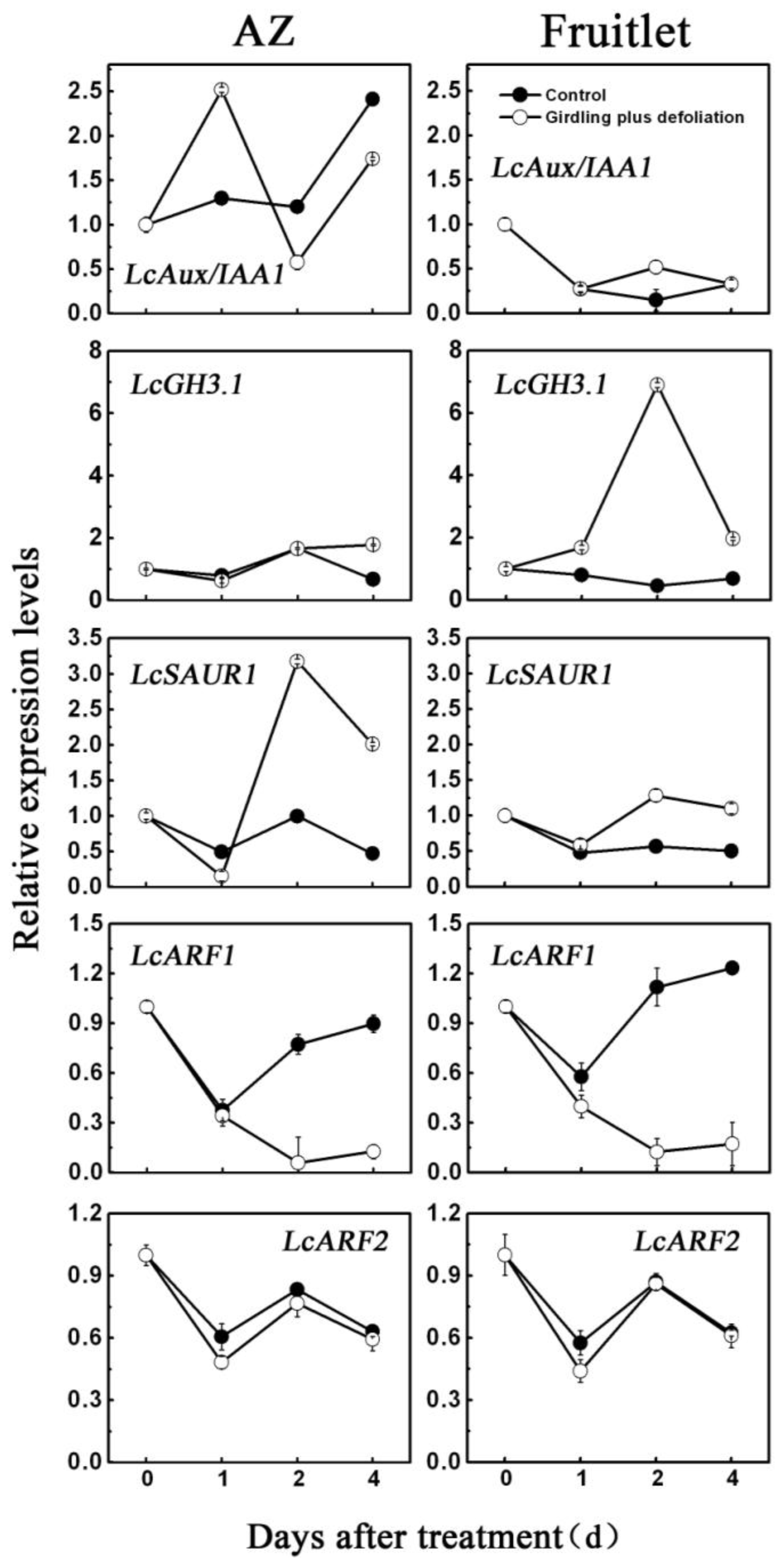
2.3. Effect of Girdling Plus Defoliation on the Expression of Litchi AUX/IAA, GH3, SAUR and ARF Genes
3. Discussion
4. Experimental Sections
4.1. Plant Materials and Treatment
4.2. RNA Isolation and cDNA Cloning
4.3. Abscission Rate Evaluation
4.4. Measurement of Endogenous IAA Content
4.5. Quantitative Real-Time PCR Analysis
5. Conclusions
Acknowledgements
- Conflict of InterestThe authors declare no conflict of interest.
References
- Sun, L.; Bukovac, M.J.; Forsline, P.L.; Van Nocker, S. Natural variation in fruit abscission-related traits in apple (Malus). Euphytica 2009, 165, 55–67. [Google Scholar]
- Ish-Shalom, M.; Dahan, Y.; Maayan, I.; Irihimovitch, V. Cloning and molecular characterization of an ethylene receptor gene, MiERS1, expressed during mango fruitlet abscission and fruit ripening. Plant Physiol. Biochem 2011, 49, 931–936. [Google Scholar]
- Yuan, R.; Kender, W.J.; Burns, J.K. Young fruit and auxin transport inhibitors affect the response of mature “Valencia” oranges to abscission materials via changing endogenous plant hormones. J. Am. Soc. Hort. Sci 2003, 128, 302–308. [Google Scholar]
- Bonghi, C.; Tonutti, P.; Ramina, A. Biochemical and molecular aspects of fruitlet abscission. Plant Growth Regul 2000, 31, 35–42. [Google Scholar]
- De Almeida, I.M.L.; Rodrigues, J.D.; Ono, E.O. Application of plant growth regulators at pre-harvest for fruit development of “Pera” oranges. Braz. Arch. Biol. Technol 2004, 47, 511–520. [Google Scholar]
- Abel, S.; Theologis, A. Early genes and auxin action. Plant Physiol 1996, 111, 9–17. [Google Scholar]
- Hagen, G.; Guilfoyle, T. Auxin-Responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol 2002, 49, 373–385. [Google Scholar]
- Dharmasiri, N.; Dharmasiri, S.; Jones, A.M.; Estelle, M. Auxin action in a cell free system. Curr. Biol 2003, 13, 1418–1422. [Google Scholar]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar]
- Worley, C.K.; Zenser, N.; Ramos, J.; Rouse, D.; Leyser, O.; Theologis, A.; Callis, J. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 2000, 21, 553–562. [Google Scholar]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inze, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 2012, 70, 978–990. [Google Scholar]
- Chae, K.; Isaacs, C.G.; Reeves, P.H.; Maloney, G.S.; Muday, G.K.; Nagpal, P.; Reed, J.W. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyls and stamen filament elongation. Plant J 2012, 71, 684–691. [Google Scholar]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2004, 15, 533–543. [Google Scholar]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 1998, 17, 1405–1411. [Google Scholar]
- Harper, R.M.; Stowe-Evans, E.L.; Luesse, D.R.; Muto, H.; Tatematsu, K.; Watahiki, M.K.; Yamamoto, K.; Liscum, E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 2000, 12, 757–770. [Google Scholar]
- Abebie, B.; Lers, A.; Philosoph-Hadas, S.; Goren, R.; Riov, J.; Meir, S. Differential effects of NAA and 2,4-D in reducing floret abscission in cestrum (Cestrum elegans Schlecht) cut flowers are associated with their differential activation of Aux/IAA homologous genes. Ann. Bot 2008, 101, 249–259. [Google Scholar]
- Abebie, B.; Goren, R.; Huberman, M.; Meir, S.; Philosoph-Hadas, S.; Riov, J. Prevention of bud and floret abscission in Cestrum cut flowers is related to the mode of transport and metabolism of synthetic auxins. Acta Hortic 2005, 682, 789–794. [Google Scholar]
- Meir, S.; Hunter, D.A.; Chen, J.C.; Halaly, V.; Reid, M.S. Molecular changes occurring during acquisition of abscission competence following auxin depletion in Mirabilis jalapa. Plant Physiol 2006, 141, 1604–1616. [Google Scholar]
- Zuo, X.; Xu, T.; Qi, M.; Lv, S.; Li, J.; Gao, S.; Li, T. Expression patterns of auxin-responsive genes during tomato flower pedicel abscission and potential effects of calcium. Aust. J. Bot 2012, 60, 68–78. [Google Scholar]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar]
- Jiang, Y.M.; Chen, F. A study on polyamine change and browning of fruit during cold storage of litchi (Litchi chinensis Sonn.). Postharvest Biol. Tec 1995, 5, 245–250. [Google Scholar]
- Mitra, S.K.; Pereira, L.S.; Pathak, P.K.; Majumadar, D. Fruit abscission pattern of lychee cultivars. Acta Hortic 2005, 665, 215–218. [Google Scholar]
- Stern, R.A.; Kigel, J.; Tomer, E.; Gazit, S. “Mauritius” lychee fruit development and Rreduced abscission after treatment with the auxin 2,4,5-TP. J. Am. Soc. Hort. Sci 1995, 120, 65–70. [Google Scholar]
- Yuan, R.; Huang, H. Litchi fruit abscission: Its patterns, effect of shading and relation to endogenous abscisic acid. Scientia Hort 1988, 36, 281–292. [Google Scholar]
- Rungruchkanont, K.; Ketsa, S.; Chatchawankanphanich, O.; Van Doorn, W.G. Endogenous auxin regulates the sensitivity of Dendrobium (cv. Miss Ten) flower pedicel abscission to ethylene. Funct. Plant Biol 2007, 34, 885–894. [Google Scholar]
- Meir, S.; Philosoph-Hadas, S.; Sundaresan, S.; Selvaraj, K.S.; Burd, S.; Ophir, R.; Kochanek, B.; Reid, M.S.; Jiang, C.Z.; Lers, A. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 2010, 154, 1929–1956. [Google Scholar]
- Agusti, M.; Almela, V.; Juan, M.; Alferez, F.; Tadeo, F.R.; Zacarias, L. Histological and physiological characterization of rind breakdown of “Navelate” sweet orange. Ann. Bot 2001, 88, 415–422. [Google Scholar]
- Stern, R.A.; Gazit, S. The reproductive biology of the lychee. Hortic Rev 2003, 28, 393–453. [Google Scholar]
- Yuan, R.; Huang, H. Effect of NAA, NAA plus nucleotides on fruit set of lychee. Austral. Lychee Yearbook 1991, 1, 46–50. [Google Scholar]
- Obeso, J.R. Effects of defoliation and girdling on fruit production in Ilex aquifolium. Funct. Ecol 1998, 12, 486–491. [Google Scholar]
- Gómez-Cadenas, A.; Mehouachi, J.; Tadeo, F.R.; Primo-Millo, E.; Talon, M. Hormonal regulation of fruitlet abscission induced by carbohydrate shortage in citrus. Planta 2000, 210, 636–643. [Google Scholar]
- Liu, D.J.; Chen, J.Y.; Lu, W.J. Expression and regulation of the early auxin-responsive Aux/IAA genes during strawberry fruit development. Mol. Biol. Rep 2011, 38, 1187–1193. [Google Scholar]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar]
- Terol, J.; Domingo, C.; Talon, M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene 2006, 371, 279–290. [Google Scholar]
- Kuang, J.F.; Zhang, Y.; Chen, J.Y.; Chen, Q.J.; Jiang, Y.M.; Lin, H.T.; Xu, S.J.; Lu, W.J. Two GH3 genes from longan are differentially regulated during fruit growth and development. Gene 2011, 485, 1–6. [Google Scholar]
- Park, J.E.; Kim, Y.S.; Yoon, H.K.; Park, C.M. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci 2007, 172, 150–157. [Google Scholar]
- Reddy, V.S.; Ali, G.S.; Reddy, A.S. Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem 2002, 277, 9840–9852. [Google Scholar]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar]
- Ruperti, B.; Cattivelli, L.; Pagni, S.; Ramina, A. Ethylene-Responsive genes are differentially regulated during abscission, organ senescence and wounding in peach (Prunus persica). J. Exp. Bot 2002, 53, 429–437. [Google Scholar]
- Stern, R.A.; Meron, M.; Naor, A.; Wallach, R.; Bravdo, B.; Gazit, S. Effect of fall irrigation level in “Mauritius” and “Floridian” lychee on soil and plant water status, flowering intensity, and yield. J. Am. Soc. Hort. Sci 1998, 123, 150–155. [Google Scholar]
- Gillaspy, G.; Ben-David, H.; Gruissem, W. Fruits: A develop-mental perspective. Plant Cell 1993, 5, 1439–1451. [Google Scholar]
- Roberts, J.A.; Whitelaw, C.A.; Gonzales-Carranza, Z.H.; McManus, M.T. Cell separation processes in plants—models, mechanisms and manipulation. Ann. Bot 2000, 86, 223–235. [Google Scholar]
- Ouellet, F.; Overvoorde, P.J.; Theologis, A. A IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 2001, 13, 829–841. [Google Scholar]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-Wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar]
- Zhou, L.; Jan, J.C.; Jones, T.L.; Sheen, J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. USA 1998, 95, 10294–10299. [Google Scholar]
- Weijers, D.; Benkova, E.; Jager, K.E.; Schlereth, A.; Hamann, T.; Kientz, M.; Wilmoth, J.C.; Reed, J.W.; Jurgens, G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 2005, 24, 1874–1885. [Google Scholar]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol 2002, 49, 387–400. [Google Scholar]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar]
- Wan, C.Y.; Wilkins, T.A. A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem 1994, 223, 7–12. [Google Scholar]
- Wu, J.Y.; Peng, G.; Li, C.Q.; Lu, W.J.; Wang, Z.H.; Li, J.G. A new rapid and effective method for RNA isolation from litchi tissues of fruitlet and abscission zone. Acta Hortic. Sin 2011, 38, 1191–1196. [Google Scholar]
- Lin, W.H.; Wang, Y.; Mueller-Roeber, B.; Brearley, C.A.; Xu, Z.H.; Xue, H.W. At5PTase13 modulates cotyledon vein development through regulating auxin homeostasis. Plant Physiol 2005, 139, 1677–1691. [Google Scholar]
- Chen, J.G.; Wang, S.; Lazarus, C.M.; Napier, R.M.; Jones, J.M. Altered expression of Auxin-binding Protein 1 affects cell expansion and auxin pool size in tobacco cells. J. Plant Growth Regul 2006, 25, 69–78. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar]
- Zhong, H.Y.; Chen, J.W.; Li, C.Q.; Chen, L.; Wu, J.Y.; Chen, J.Y.; Lu, W.J.; Li, J.G. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep. 2011, 30, 641–653. [Google Scholar]
| Name | Sequences (5′-3′) |
|---|---|
| AUX/IAA-For | GGTGGTGCGCTGGCGNCCNRT |
| AUX/IAA-Rev | CGATCGCCTCGGACCGYTTNATDAT |
| GH3-For | CTACGACACCTACCARCAGWTSTAYWSNCA |
| GH3-Rev | CAGCTCCCAGAAGATCAYGTRRTSNCCNGG |
| SAUR-5RACE1 | AGGATGTACTCGAAGGTTGCCWYNTCRCANGG |
| SAUR-5RACE2 | CCTCCTGCTCGAAGCCGWAYTCYTCNKC |
| ARF-For | TGGAGGTTCAGGCACATCTTCMGNGGNCARCC |
| ARF-Rev | AGCAGAACTCCTCCCATGGRTCRTCNCC |
| LcAUX/IAA1-3RACE1 | GTAAAGGTGGCTGTGGATGG |
| LcAUX/IAA1-3RACE2 | TGAGGAGATGTTTTCGTGCT |
| LcAUX/IAA1-5RACE1 | GTCGCCATCATTGTCCTCATAAG |
| LcAUX/IAA1-5RACE2 | ACATCTCCTCAAGAGCAGTCAAG |
| LcGH3.1-3RACE1 | CGACCCTACGACCCTTACAA |
| LcGH3.1-3RACE2 | CCTACAAGTCCTCCGTCTCG |
| LcGH3.1-5RACE1 | CACAGAAGGGTCAGTGATTTGGG |
| LcGH3.1-5RACE2 | CCAGAGGCAAAAATAGCACCAAC |
| LcSAUR1-3RACE1 | GCAGGAGGTTCATCGTTCG |
| LcSAUR1-3RACE2 | GCACCTGAACCACCCTCTTT |
| LcARF1-3RACE1 | CGAACAAGGAAGTGGATGCT |
| LcARF1-3RACE2 | GCAACCTCTGAATCCGCAAG |
| LcARF1-5RACE1 | TTGCCCAACTCTGCTGGAAGGTA |
| LcARF1-5RACE2 | GAGCATCCACTTCCTTGTTCGTT |
| LcARF2-3RACE1 | ACCCAACTACCCCAACCTTC |
| LcARF2-3RACE2 | ACGGATGAGGTCTACGCACA |
| LcARF2-5RACE1 | TGTGCGTAGACCTCATCCGTTTC |
| LcARF2-5RACE2 | CTTGGAAGGAAGGTTGGGGTAGT |
| Name | Forward primer(5′-3′) | Reverse primer(5′-3′) |
|---|---|---|
| LcAUX/IAA1 | TGAAGCAACAATAAGTGGAAAAGG | TTCGCGTGAAACAAGATGTTGTA |
| LcGH3.1 | GCCACACTGCTGATAGAGAGACT | TCCCCTCCTGATGTCCCAGAACT |
| LcSAUR1 | AGTGCAACAAGATTCGTCACATTG | GCGCTGCCGATGACCCTCTAGAC |
| LcARF1 | CAAAGAAGAAGTGGAGAAGATGAC | ATGTAACTAGGTTCAGGTTTTCAC |
| LcARF2 | AGGGGTTGAATCCTTAAATCCAAG | ATTTTCGTGGGATTATGTTATGTC |
| LcACTIN | ACCGTATGAGCAAGGAAATCACTG | TCGTCGTACTCACCCTTTGAAATC |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kuang, J.-F.; Wu, J.-Y.; Zhong, H.-Y.; Li, C.-Q.; Chen, J.-Y.; Lu, W.-J.; Li, J.-G. Carbohydrate Stress Affecting Fruitlet Abscission and Expression of Genes Related to Auxin Signal Transduction Pathway in Litchi. Int. J. Mol. Sci. 2012, 13, 16084-16103. https://doi.org/10.3390/ijms131216084
Kuang J-F, Wu J-Y, Zhong H-Y, Li C-Q, Chen J-Y, Lu W-J, Li J-G. Carbohydrate Stress Affecting Fruitlet Abscission and Expression of Genes Related to Auxin Signal Transduction Pathway in Litchi. International Journal of Molecular Sciences. 2012; 13(12):16084-16103. https://doi.org/10.3390/ijms131216084
Chicago/Turabian StyleKuang, Jian-Fei, Jian-Yang Wu, Hai-Ying Zhong, Cai-Qin Li, Jian-Ye Chen, Wang-Jin Lu, and Jian-Guo Li. 2012. "Carbohydrate Stress Affecting Fruitlet Abscission and Expression of Genes Related to Auxin Signal Transduction Pathway in Litchi" International Journal of Molecular Sciences 13, no. 12: 16084-16103. https://doi.org/10.3390/ijms131216084





