Antibody Phage Display Libraries: Contributions to Oncology
Abstract
:1. Monoclonal Antibodies, Useful Diagnostic and Therapeutic Molecules
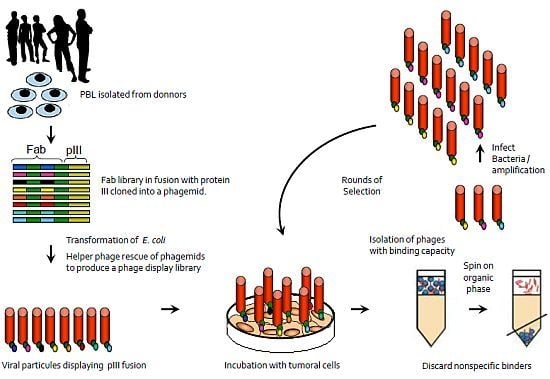
2. Antibody Phage Display Libraries
2.1. Immunized Phage Display Libraries
2.2. Immunized Animals as Source of Antibody Repertoire
2.3. Nom-Immune Libraries to Identify Binders to Tumor Markers
3. Phage Display to Select Antibodies for Molecular Imaging
4. Antibodies Derived from Phage Display Libraries Used in Clinical Trials
4.1. 1D09C3 (GPC Biotech AG)
4.2. Fresolimumab (GC-1008—Cambridge Antibody Technology/Genzyme)
4.3. Cixutumumab (IMC-A12—ImClone Systems Incorporated)
4.4. Necitumumab (IMC-11F8—ImClone Systems Incorporated)
4.5. Ramucirumab (IMC-1121B—ImClone Systems Incorporated)
4.6. Mapatumumab (HGS-ETR1—Human Genome Sciences Inc./GlaxoSmithKline)
4.7. Lexatumumab (HGS-ETR2—Human Genome Sciences Inc.)
4.8. Moxetumomab Pasudotox (HA22, CAT-8015, (RFB4(dsFv)-PE38—MedImmune)
5. Conclusions
References
- Berger, M.; Shankar, V.; Vafai, A. Therapeutic applications of monoclonal antibodies. Am. J. Med. Sci 2002, 324, 14–30. [Google Scholar]
- Seymour, J.F. New treatment approaches to indolent non-Hodgkin’s lymphoma. Semin. Oncol 2004, 31, 27–32. [Google Scholar]
- Cosimi, A.B.; Colvin, R.B.; Burton, R.C.; Rubin, R.H.; Goldstein, G.; Kung, P.C.; Hansen, W.P.; Delmonico, F.L.; Russell, P.S. Use of monoclonal antibodies to T-cell subsets for immunologic monitoring and treatment in recipients of renal allografts. N. Engl. J. Med 1981, 305, 308–314. [Google Scholar]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar]
- Maranhao, A.Q.; Brigido, M.M. Expression of anti-Z-DNA single chain antibody variable fragment on the filamentous phage surface. Braz. J. Med. Biol. Res 2000, 33, 569–579. [Google Scholar]
- Morrison, S.L.; Oi, V.T. Genetically engineered antibody molecules. Adv. Immunol 1989, 44, 65–92. [Google Scholar]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol 2005, 23, 1126–1136. [Google Scholar]
- Holt, L.J.; Herring, C.; Jespers, L.S.; Woolven, B.P.; Tomlinson, I.M. Domain antibodies: Proteins for therapy. Trend. Biotechnol 2003, 21, 484–490. [Google Scholar]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar]
- Thie, H.; Meyer, T.; Schirrmann, T.; Hust, M.; Dubel, S. Phage display derived therapeutic antibodies. Curr. Pharm. Biotechnol 2008, 9, 439–446. [Google Scholar]
- ClinicalTrials.gov Home Page. Available online: http://clinicaltrials.gov accessed on 3 May 2012.
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar]
- Posner, B.; Smiley, J.; Lee, I.; Benkovic, S. Catalytic antibodies: Perusing combinatorial libraries. Trends Biochem. Sci 1994, 19, 145–150. [Google Scholar]
- Barbas, C.F., 3rd; Kang, A.S.; Lerner, R.A.; Benkovic, S.J. Assembly of combinatorial antibody libraries on phage surfaces: The gene III site. Proc. Natl. Acad. Sci. USA 1991, 88, 7978–7982. [Google Scholar]
- Clackson, T.; Hoogenboom, H.R.; Griffiths, A.D.; Winter, G. Making antibody fragments using phage display libraries. Nature 1991, 352, 624–628. [Google Scholar]
- Tomlinson Library Home Page. Available online: http://www.coloradomesa.edu/cmulibrary/index.html accessed on 3 April 2012.
- Chinestra, P.; Lajoie-Mazenc, I.; Faye, J.C.; Favre, G. Use of phage display for the identification of molecular sensors specific for activated Rho. Meth. Mol. Biol 2012, 827, 283–303. [Google Scholar]
- Davern, S.M.; Foote, L.J.; Lankford, T.K.; Macy, S.D.; Wall, M.D.; Kennel, S.J. Identification of an antilaminin-1 scFv that preferentially homes to vascular solid tumors. Cancer Biother. Radiopharm 2005, 20, 524–533. [Google Scholar]
- Barbas, C.F., 3rd; Burton, D.R. Selection and evolution of high-affinity human anti-viral antibodies. Trends Biotechnol 1996, 14, 230–234. [Google Scholar]
- Dantas-Barbosa, C.; Brigido, M.M.; Maranhao, A.Q. Construction of a human Fab phage display library from antibody repertoires of osteosarcoma patients. Genet. Mol. Res 2005, 4, 126–140. [Google Scholar]
- Portolano, S.; McLachlan, S.M.; Rapoport, B. High affinity, thyroid-specific human autoantibodies displayed on the surface of filamentous phage use V genes similar to other autoantibodies. J. Immunol 1993, 151, 2839–2851. [Google Scholar]
- Griffiths, A.D.; Malmqvist, M.; Marks, J.D.; Bye, J.M.; Embleton, M.J.; McCafferty, J.; Baier, M.; Holliger, K.P.; Gorick, B.D.; Hughes-Jones, N.C.; et al. Human anti-self antibodies with high specificity from phage display libraries. EMBO J 1993, 12, 725–734. [Google Scholar]
- Mandecki, W.; Chen, Y.C.; Grihalde, N. A mathematical model for biopanning (affinity selection) using peptide libraries on filamentous phage. J. Theor. Biol 1995, 176, 523–530. [Google Scholar]
- Marks, J.D.; Hoogenboom, H.R.; Bonnert, T.P.; McCafferty, J.; Griffiths, A.D.; Winter, G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol 1991, 222, 581–597. [Google Scholar]
- Vaughan, T.J.; Williams, A.J.; Pritchard, K.; Osbourn, J.K.; Pope, A.R.; Earnshaw, J.C.; McCafferty, J.; Hodits, R.A.; Wilton, J.; Johnson, K.S. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol 1996, 14, 309–314. [Google Scholar]
- Pasqualini, R.; Ruoslahti, E. Organ targeting in vivo using phage display peptide libraries. Nature 1996, 380, 364–366. [Google Scholar]
- Glokler, J.; Schutze, T.; Konthur, Z. Automation in the high-throughput selection of random combinatorial libraries—Different approaches for select applications. Molecules 2010, 15, 2478–2490. [Google Scholar]
- Williams, B.R.; Sharon, J. Polyclonal anti-colorectal cancer Fab phage display library selected in one round using density gradient centrifugation to separate antigen-bound and free phage. Immunol. Lett 2002, 81, 141–148. [Google Scholar]
- Giordano, R.J.; Cardo-Vila, M.; Lahdenranta, J.; Pasqualini, R.; Arap, W. Biopanning and rapid analysis of selective interactive ligands. Nat. Med 2001, 7, 1249–1253. [Google Scholar]
- Liu, Y.; Adams, J.D.; Turner, K.; Cochran, F.V.; Gambhir, S.S.; Soh, H.T. Controlling the selection stringency of phage display using a microfluidic device. Lab Chip 2009, 9, 1033–1036. [Google Scholar]
- He, M.; Taussig, M.J. Antibody-ribosome-mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res 1997, 25, 5132–5134. [Google Scholar]
- Wang, X.X.; Shusta, E.V. The use of scFv-displaying yeast in mammalian cell surface selections. J. Immunol. Meth 2005, 304, 30–42. [Google Scholar]
- Amstutz, P.; Forrer, P.; Zahnd, C.; Pluckthun, A. In vitro display technologies: Novel developments and applications. Curr. Opin. Biotechnol 2001, 12, 400–405. [Google Scholar]
- Michelfelder, S.; Lee, M.K.; deLima-Hahn, E.; Wilmes, T.; Kaul, F.; Muller, O.; Kleinschmidt, J.A.; Trepel, M. Vectors selected from adeno-associated viral display peptide libraries for leukemia cell-targeted cytotoxic gene therapy. Exp. Hematol 2007, 35, 1766–1776. [Google Scholar]
- Grabherr, R.; Ernst, W. Baculovirus for eukaryotic protein display. Curr. Gene Ther 2010, 10, 195–200. [Google Scholar]
- Pepper, L.R.; Cho, Y.K.; Boder, E.T.; Shusta, E.V. A decade of yeast surface display technology: Where are we now? Comb. Chem. High Throughput Screen 2008, 11, 127–134. [Google Scholar]
- Skerra, A. Alternative non-antibody scaffolds for molecular recognition. Curr. Opin. Biotechnol 2007, 18, 295–304. [Google Scholar]
- Zoller, F.; Haberkorn, U.; Mier, W. Miniproteins as phage display-scaffolds for clinical applications. Molecules 2011, 16, 2467–2485. [Google Scholar]
- Wortzel, R.D.; Urban, J.L.; Philipps, C.; Fitch, F.W.; Schreiber, H. Independent immunodominant and immunorecessive tumor-specific antigens on a malignant tumor: Antigenic dissection with cytolytic T cell clones. J. Immunol 1983, 130, 2461–2466. [Google Scholar]
- Cai, X.; Garen, A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: Selection of specific antibodies from single-chain Fv fusion phage libraries. Proc. Natl. Acad. Sci. USA 1995, 92, 6537–6541. [Google Scholar]
- Pereira, S.; Maruyama, H.; Siegel, D.; van Belle, P.; Elder, D.; Curtis, P.; Herlyn, D. A model system for detection and isolation of a tumor cell surface antigen using antibody phage display. J. Immunol. Meth 1997, 203, 11–24. [Google Scholar]
- Dantas-Barbosa, C.; Faria, F.P.; Brigido, M.M.; Maranhao, A.Q. Isolation of osteosarcoma-associated human antibodies from a combinatorial Fab phage display library. J. Biomed. Biotechnol. [CrossRef]
- Figini, M.; Obici, L.; Mezzanzanica, D.; Griffiths, A.; Colnaghi, M.I.; Winter, G.; Canevari, S. Panning phage antibody libraries on cells: Isolation of human Fab fragments against ovarian carcinoma using guided selection. Cancer Res 1998, 58, 991–996. [Google Scholar]
- Lee, K.J.; Mao, S.; Sun, C.; Gao, C.; Blixt, O.; Arrues, S.; Hom, L.G.; Kaufmann, G.F.; Hoffman, T.Z.; Coyle, A.R.; et al. Phage-display selection of a human single-chain fv antibody highly specific for melanoma and breast cancer cells using a chemoenzymatically synthesized G(M3)-carbohydrate antigen. J. Am. Chem. Soc 2002, 124, 12439–12446. [Google Scholar]
- Sahin, U.; Tureci, O.; Schmitt, H.; Cochlovius, B.; Johannes, T.; Schmits, R.; Stenner, F.; Luo, G.; Schobert, I.; Pfreundschuh, M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc. Natl. Acad. Sci. USA 1995, 92, 11810–11813. [Google Scholar]
- Scanlan, M.J.; Gordan, J.D.; Williamson, B.; Stockert, E.; Bander, N.H.; Jongeneel, V.; Gure, A.O.; Jager, D.; Jager, E.; Knuth, A.; et al. Antigens recognized by autologous antibody in patients with renal-cell carcinoma. Int. J. Cancer 1999, 83, 456–464. [Google Scholar]
- Hufton, S.E.; Moerkerk, P.; de Bruine, A.; Arends, J.W.; Hoogenboom, H.R. Serological antigen selection of phage displayed colorectal tumour cDNA libraries. Biochem. Soc.Trans 1998, 26, S5. [Google Scholar]
- Somers, V.A.; Brandwijk, R.J.; Joosten, B.; Moerkerk, P.T.; Arends, J.W.; Menheere, P.; Pieterse, W.O.; Claessen, A.; Scheper, R.J.; Hoogenboom, H.R.; et al. A panel of candidate tumor antigens in colorectal cancer revealed by the serological selection of a phage displayed cDNA expression library. J. Immunol 2002, 169, 2772–2780. [Google Scholar]
- Fernandez-Madrid, F.; Tang, N.; Alansari, H.; Granda, J.L.; Tait, L.; Amirikia, K.C.; Moroianu, M.; Wang, X.; Karvonen, R.L. Autoantibodies to annexin XI-A and other autoantigens in the diagnosis of breast cancer. Cancer Res 2004, 64, 5089–5096. [Google Scholar]
- Wang, X.; Yu, J.; Sreekumar, A.; Varambally, S.; Shen, R.; Giacherio, D.; Mehra, R.; Montie, J.E.; Pienta, K.J.; Sanda, M.G.; et al. Autoantibody signatures in prostate cancer. N. Engl. J. Med 2005, 353, 1224–1235. [Google Scholar]
- Chen, G.; Wang, X.; Yu, J.; Varambally, S.; Yu, J.; Thomas, D.G.; Lin, M.Y.; Vishnu, P.; Wang, Z.; Wang, R.; et al. Autoantibody profiles reveal ubiquilin 1 as a humoral immune response target in lung adenocarcinoma. Cancer Res 2007, 67, 3461–3467. [Google Scholar]
- Lee, S.Y.; Obata, Y.; Yoshida, M.; Stockert, E.; Williamson, B.; Jungbluth, A.A.; Chen, Y.T.; Old, L.J.; Scanlan, M.J. Immunomic analysis of human sarcoma. Proc. Natl. Acad. Sci. USA 2003, 100, 2651–2656. [Google Scholar]
- McWhirter, J.R.; Kretz-Rommel, A.; Saven, A.; Maruyama, T.; Potter, K.N.; Mockridge, C.I.; Ravey, E.P.; Qin, F.; Bowdish, K.S. Antibodies selected from combinatorial libraries block a tumor antigen that plays a key role in immunomodulation. Proc. Natl. Acad. Sci. USA 2006, 103, 1041–1046. [Google Scholar]
- Yang, J.; Baskar, S.; Kwong, K.Y.; Kennedy, M.G.; Wiestner, A.; Rader, C. Therapeutic potential and challenges of targeting receptor tyrosine kinase ROR1 with monoclonal antibodies in B-cell malignancies. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Stein, C.; Kellner, C.; Kugler, M.; Reiff, N.; Mentz, K.; Schwenkert, M.; Stockmeyer, B.; Mackensen, A.; Fey, G.H. Novel conjugates of single-chain Fv antibody fragments specific for stem cell antigen CD123 mediate potent death of acute myeloid leukaemia cells. Br. J. Haematol 2010, 148, 879–889. [Google Scholar]
- Baral, T.N.; Murad, Y.; Nguyen, T.D.; Iqbal, U.; Zhang, J. Isolation of functional single domain antibody by whole cell immunization: Implications for cancer treatment. J. Immunol. Meth 2011, 371, 70–80. [Google Scholar]
- Bell, A.; Wang, Z.J.; Arbabi-Ghahroudi, M.; Chang, T.A.; Durocher, Y.; Trojahn, U.; Baardsnes, J.; Jaramillo, M.L.; Li, S.; Baral, T.N.; et al. Differential tumor-targeting abilities of three single-domain antibody formats. Cancer Lett 2010, 289, 81–90. [Google Scholar] [Green Version]
- Klagsbrun, M.; D’Amore, P.A. Vascular endothelial growth factor and its receptors. Cytokine Growth Factor Rev 1996, 7, 259–270. [Google Scholar]
- Kastelic, D.; Frkovic-Grazio, S.; Baty, D.; Truan, G.; Komel, R.; Pompon, D. A single-step procedure of recombinant library construction for the selection of efficiently produced llama VH binders directed against cancer markers. J. Immunol. Meth 2009, 350, 54–62. [Google Scholar]
- Huang, Y.J.; Chen, I.C.; Yu, C.M.; Lee, Y.C.; Hsu, H.J.; Ching, A.T.; Chang, H.J.; Yang, A.S. Engineering anti-vascular endothelial growth factor single chain disulfide-stabilized antibody variable fragments (sc-dsFv) with phage-displayed sc-dsFv libraries. J. Biol. Chem 2010, 285, 7880–7891. [Google Scholar]
- Wang, X.; Zhong, P.; Luo, P.P.; Wang, K.C. Antibody engineering using phage display with a coiled-coil heterodimeric Fv antibody fragment. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Lu, D.; Jimenez, X.; Zhang, H.; Bohlen, P.; Witte, L.; Zhu, Z. Selection of high affinity human neutralizing antibodies to VEGFR2 from a large antibody phage display library for antiangiogenesis therapy. Int. J. Cancer 2002, 97, 393–399. [Google Scholar]
- Hetian, L.; Ping, A.; Shumei, S.; Xiaoying, L.; Luowen, H.; Jian, W.; Lin, M.; Meisheng, L.; Junshan, Y.; Chengchao, S. A novel peptide isolated from a phage display library inhibits tumor growth and metastasis by blocking the binding of vascular endothelial growth factor to its kinase domain receptor. J. Biol. Chem 2002, 277, 43137–43142. [Google Scholar]
- Lin, Z.; Cao, P.; Lei, H. Identification of a neutralizing scFv binding to human vascular endothelial growth factor 165 (VEGF165) using a phage display antibody library. Appl. Biochem. Biotechnol 2008, 144, 15–26. [Google Scholar]
- Lamdan, H.; Ayala, M.; Rojas, G.; Munoz, Y.; Morera, Y.; Guirola, O.; Chinea, G.; Gavilondo, J.V. Isolation of a novel neutralizing antibody fragment against human vascular endothelial growth factor from a phage-displayed human antibody repertoire using an epitope disturbing strategy. J. Biotechnol 2011, 151, 166–174. [Google Scholar]
- Rinderknecht, M.; Villa, A.; Ballmer-Hofer, K.; Neri, D.; Detmar, M. Phage-derived fully human monoclonal antibody fragments to human vascular endothelial growth factor-C block its interaction with VEGF receptor-2 and 3. PLoS One 2010, 5. [Google Scholar] [CrossRef]
- Chang, D.K.; Chiu, C.Y.; Kuo, S.Y.; Lin, W.C.; Lo, A.; Wang, Y.P.; Li, P.C.; Wu, H.C. Antiangiogenic targeting liposomes increase therapeutic efficacy for solid tumors. J. Biol. Chem 2009, 284, 12905–12916. [Google Scholar]
- Carpenter, G. Receptors for epidermal growth factor and other polypeptide mitogens. Annu. Rev. Biochem 1987, 56, 881–914. [Google Scholar]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar]
- Yip, Y.L.; Smith, G.; Koch, J.; Dubel, S.; Ward, R.L. Identification of epitope regions recognized by tumor inhibitory and stimulatory anti-ErbB-2 monoclonal antibodies: Implications for vaccine design. J. Immunol 2001, 166, 5271–5278. [Google Scholar]
- Jasinska, J.; Wagner, S.; Radauer, C.; Sedivy, R.; Brodowicz, T.; Wiltschke, C.; Breiteneder, H.; Pehamberger, H.; Scheiner, O.; Wiedermann, U.; et al. Inhibition of tumor cell growth by antibodies induced after vaccination with peptides derived from the extracellular domain of Her-2/neu. Int. J. Cancer 2003, 107, 976–983. [Google Scholar]
- Alvarez-Rueda, N.; Ladjemi, M.Z.; Behar, G.; Corgnac, S.; Pugniere, M.; Roquet, F.; Bascoul-Mollevi, C.; Baty, D.; Pelegrin, A.; Navarro-Teulon, I. A llama single domain anti-idiotypic antibody mimicking HER2 as a vaccine: Immunogenicity and efficacy. Vaccine 2009, 27, 4826–4833. [Google Scholar]
- Coelho, M.; Gauthier, P.; Pugniere, M.; Roquet, F.; Pelegrin, A.; Navarro-Teulon, I. Isolation and characterisation of a human anti-idiotypic scFv used as a surrogate tumour antigen to elicit an anti-HER-2/neu humoral response in mice. Br. J. Cancer 2004, 90, 2032–2041. [Google Scholar]
- Zehnder-Fjallman, A.H.; Marty, C.; Halin, C.; Hohn, A.; Schibli, R.; Ballmer-Hofer, K.; Schwendener, R.A. Evaluation of anti-VEGFR-3 specific scFv antibodies as potential therapeutic and diagnostic tools for tumor lymph-angiogenesis. Oncol. Rep 2007, 18, 933–941. [Google Scholar]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci 2002, 115, 3861–3863. [Google Scholar]
- Neri, D.; Carnemolla, B.; Nissim, A.; Leprini, A.; Querze, G.; Balza, E.; Pini, A.; Tarli, L.; Halin, C.; Neri, P.; et al. Targeting by affinity-matured recombinant antibody fragments of an angiogenesis associated fibronectin isoform. Nat. Biotechnol 1997, 15, 1271–1275. [Google Scholar]
- Rossin, R.; Berndorff, D.; Friebe, M.; Dinkelborg, L.M.; Welch, M.J. Small-animal PET of tumor angiogenesis using a (76)Br-labeled human recombinant antibody fragment to the ED-B domain of fibronectin. J. Nucl. Med 2007, 48, 1172–1179. [Google Scholar]
- Tijink, B.M.; Perk, L.R.; Budde, M.; Stigter-van Walsum, M.; Visser, G.W.; Kloet, R.W.; Dinkelborg, L.M.; Leemans, C.R.; Neri, D.; van Dongen, G.A. (124)I-L19-SIP for immuno-PET imaging of tumour vasculature and guidance of (131)I-L19-SIP radioimmunotherapy. Eur. J. Nucl. Med. Mol. Imag 2009, 36, 1235–1244. [Google Scholar]
- Valadon, P.; Garnett, J.D.; Testa, J.E.; Bauerle, M.; Oh, P.; Schnitzer, J.E. Screening phage display libraries for organ-specific vascular immunotargeting in vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 407–412. [Google Scholar]
- Lu, R.M.; Chang, Y.L.; Chen, M.S.; Wu, H.C. Single chain anti-c-Met antibody conjugated nanoparticles for in vivo tumor-targeted imaging and drug delivery. Biomaterials 2011, 32, 3265–3274. [Google Scholar]
- Malviya, G.; de Vries, E.F.; Dierckx, R.A.; Signore, A. Synthesis and evaluation of 99mTc-labelled monoclonal antibody 1D09C3 for molecular imaging of major histocompatibility complex class II protein expression. Mol. Imag. Biol 2011, 13, 930–939. [Google Scholar]
- Carlo-Stella, C.; Di Nicola, M.; Turco, M.C.; Cleris, L.; Lavazza, C.; Longoni, P.; Milanesi, M.; Magni, M.; Ammirante, M.; Leone, A.; et al. The anti-human leukocyte antigen-DR monoclonal antibody 1D09C3 activates the mitochondrial cell death pathway and exerts a potent antitumor activity in lymphoma-bearing nonobese diabetic/severe combined immunodeficient mice. Cancer Res 2006, 66, 1799–1808. [Google Scholar]
- Carlo-Stella, C.; Guidetti, A.; Di Nicola, M.; Lavazza, C.; Cleris, L.; Sia, D.; Longoni, P.; Milanesi, M.; Magni, M.; Nagy, Z.; et al. IFN-gamma enhances the antimyeloma activity of the fully human anti-human leukocyte antigen-DR monoclonal antibody 1D09C3. Cancer Res 2007, 67, 3269–3275. [Google Scholar]
- Oude Munnink, T.H.; Arjaans, M.E.; Timmer-Bosscha, H.; Schroder, C.P.; Hesselink, J.W.; Vedelaar, S.R.; Walenkamp, A.M.; Reiss, M.; Gregory, R.C.; Lub-de Hooge, M.N.; et al. PET with the 89Zr-labeled transforming growth factor-beta antibody fresolimumab in tumor models. J. Nucl. Med 2011, 52, 2001–2008. [Google Scholar]
- Burtrum, D.; Zhu, Z.; Lu, D.; Anderson, D.M.; Prewett, M.; Pereira, D.S.; Bassi, R.; Abdullah, R.; Hooper, A.T.; Koo, H.; et al. A fully human monoclonal antibody to the insulin-like growth factor I receptor blocks ligand-dependent signaling and inhibits human tumor growth in vivo. Cancer Res 2003, 63, 8912–8921. [Google Scholar]
- Rowinsky, E.K.; Youssoufian, H.; Tonra, J.R.; Solomon, P.; Burtrum, D.; Ludwig, D.L. IMC-A12, a human IgG1 monoclonal antibody to the insulin-like growth factor I receptor. Clin. Cancer Res 2007, 13, 5549s–5555s. [Google Scholar]
- Weickhardt, A.; Doebele, R.; Oton, A.; Lettieri, J.; Maxson, D.; Reynolds, M.; Brown, A.; Jackson, M.K.; Dy, G.; Adjei, A.; et al. A phase I/II study of erlotinib in combination with the anti-insulin-like growth factor-1 receptor monoclonal antibody IMC-A12 (cixutumumab) in patients with advanced non-small cell lung cancer. J. Thorac. Oncol 2012, 7, 419–426. [Google Scholar]
- Reidy, D.L.; Vakiani, E.; Fakih, M.G.; Saif, M.W.; Hecht, J.R.; Goodman-Davis, N.; Hollywood, E.; Shia, J.; Schwartz, J.; Chandrawansa, K.; et al. Randomized, phase II study of the insulin-like growth factor-1 receptor inhibitor IMC-A12, with or without cetuximab, in patients with cetuximab- or panitumumab-refractory metastatic colorectal cancer. J. Clin. Oncol 2010, 28, 4240–4246. [Google Scholar]
- Malempati, S.; Weigel, B.; Ingle, A.M.; Ahern, C.H.; Carroll, J.M.; Roberts, C.T.; Reid, J.M.; Schmechel, S.; Voss, S.D.; Cho, S.Y.; et al. Phase I/II trial and pharmacokinetic study of cixutumumab in pediatric patients with refractory solid tumors and ewing sarcoma: A report from the children’s oncology group. J. Clin. Oncol 2012, 30, 256–262. [Google Scholar]
- Vecchione, L.; Jacobs, B.; Normanno, N.; Ciardiello, F.; Tejpar, S. EGFR-targeted therapy. Exp. Cell Res 2011, 317, 2765–2771. [Google Scholar]
- Dienstmann, R.; Tabernero, J. Necitumumab, a fully human IgG1 mAb directed against the EGFR for the potential treatment of cancer. Cur. Opin. Investig. Drugs 2010, 11, 1434–1441. [Google Scholar]
- Kuenen, B.; Witteveen, P.O.; Ruijter, R.; Giaccone, G.; Dontabhaktuni, A.; Fox, F.; Katz, T.; Youssoufian, H.; Zhu, J.; Rowinsky, E.K.; et al. A phase I pharmacologic study of necitumumab (IMC-11F8), a fully human IgG1 monoclonal antibody directed against EGFR in patients with advanced solid malignancies. Clin. Cancer Res 2010, 16, 1915–1923. [Google Scholar]
- Dienstmann, R.; Felip, E. Necitumumab in the treatment of advanced non-small cell lung cancer: Translation from preclinical to clinical development. Expert Opin. Biol. Ther 2011, 11, 1223–1231. [Google Scholar]
- Ferrara, N.; Hillan, K.J.; Gerber, H.P.; Novotny, W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov 2004, 3, 391–400. [Google Scholar]
- Hsu, J.Y.; Wakelee, H.A. Monoclonal antibodies targeting vascular endothelial growth factor: Current status and future challenges in cancer therapy. BioDrugs 2009, 23, 289–304. [Google Scholar]
- Spratlin, J.L.; Cohen, R.B.; Eadens, M.; Gore, L.; Camidge, D.R.; Diab, S.; Leong, S.; O’Bryant, C.; Chow, L.Q.; Serkova, N.J.; et al. Phase I pharmacologic and biologic study of ramucirumab (IMC-1121B), a fully human immunoglobulin G1 monoclonal antibody targeting the vascular endothelial growth factor receptor-2. J. Clin. Oncol 2010, 28, 780–787. [Google Scholar]
- Mackey, J.; Gelmon, K.; Martin, M.; McCarthy, N.; Pinter, T.; Rupin, M.; Youssoufian, H. TRIO-012: A multicenter, multinational, randomized, double-blind phase III study of IMC-1121B plus docetaxel versus placebo plus docetaxel in previously untreated patients with HER2-negative, unresectable, locally recurrent or metastatic breast cancer. Clin. Breast Cancer 2009, 9, 258–261. [Google Scholar]
- Menoret, E.; Gomez-Bougie, P.; Geffroy-Luseau, A.; Daniels, S.; Moreau, P.; Le Gouill, S.; Harousseau, J.L.; Bataille, R.; Amiot, M.; Pellat-Deceunynck, C. Mcl-1L cleavage is involved in TRAIL-R1- and TRAIL-R2-mediated apoptosis induced by HGS-ETR1 and HGS-ETR2 human mAbs in myeloma cells. Blood 2006, 108, 1346–1352. [Google Scholar]
- Pukac, L.; Kanakaraj, P.; Humphreys, R.; Alderson, R.; Bloom, M.; Sung, C.; Riccobene, T.; Johnson, R.; Fiscella, M.; Mahoney, A.; et al. HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody, induces cell death in multiple tumour types in vitro and in vivo. Br. J. Cancer 2005, 92, 1430–1441. [Google Scholar]
- Duiker, E.; Dijkers, E.; Heerspink, H.L.; de Jong, S.; van der Zee, A.; Jager, P.; Kosterink, J.; de Vries, E.; Hooge, M.L. Development of a radioiodinated apoptosis-inducing ligand, rhTRAIL, and a radiolabelled agonist TRAIL receptor antibody for clinical imaging studies. Br. J. Pharmacol 2011, 165, 2203–2212. [Google Scholar]
- Younes, A.; Vose, J.M.; Zelenetz, A.D.; Smith, M.R.; Burris, H.A.; Ansell, S.M.; Klein, J.; Halpern, W.; Miceli, R.; Kumm, E.; et al. A Phase 1b/2 trial of mapatumumab in patients with relapsed/refractory non-Hodgkin’s lymphoma. Br. J. Cancer 2010, 103, 1783–1787. [Google Scholar]
- Trarbach, T.; Moehler, M.; Heinemann, V.; Kohne, C.H.; Przyborek, M.; Schulz, C.; Sneller, V.; Gallant, G.; Kanzler, S. Phase II trial of mapatumumab, a fully human agonistic monoclonal antibody that targets and activates the tumour necrosis factor apoptosis-inducing ligand receptor-1 (TRAIL-R1), in patients with refractory colorectal cancer. Br. J. Cancer 2010, 102, 506–512. [Google Scholar]
- Bouralexis, S.; Findlay, D.M.; Evdokiou, A. Death to the bad guys: Targeting cancer via Apo2L/TRAIL. Apoptosis 2005, 10, 35–51. [Google Scholar]
- Plummer, R.; Attard, G.; Pacey, S.; Li, L.; Razak, A.; Perrett, R.; Barrett, M.; Judson, I.; Kaye, S.; Fox, N.L.; et al. Phase 1 and pharmacokinetic study of lexatumumab in patients with advanced cancers. Clin. Cancer Res 2007, 13, 6187–6194. [Google Scholar]
- Wakelee, H.A.; Patnaik, A.; Sikic, B.I.; Mita, M.; Fox, N.L.; Miceli, R.; Ullrich, S.J.; Fisher, G.A.; Tolcher, A.W. Phase I and pharmacokinetic study of lexatumumab (HGS-ETR2) given every 2 weeks in patients with advanced solid tumors. Ann. Oncol 2010, 21, 376–381. [Google Scholar]
- Campana, D.; Janossy, G.; Bofill, M.; Trejdosiewicz, L.K.; Ma, D.; Hoffbrand, A.V.; Mason, D.Y.; Lebacq, A.M.; Forster, H.K. Human B cell development. I. Phenotypic differences of B lymphocytes in the bone marrow and peripheral lymphoid tissue. J. Immunol 1985, 134, 1524–1530. [Google Scholar]
- Salvatore, G.; Beers, R.; Margulies, I.; Kreitman, R.J.; Pastan, I. Improved cytotoxic activity toward cell lines and fresh leukemia cells of a mutant anti-CD22 immunotoxin obtained by antibody phage display. Clin. Cancer Res 2002, 8, 995–1002. [Google Scholar]
- Weldon, J.E.; Pastan, I. A guide to taming a toxin—Recombinant immunotoxins constructed from Pseudomonas exotoxin A for the treatment of cancer. FEBS J 2011, 278, 4683–4700. [Google Scholar]
- Alderson, R.F.; Toki, B.E.; Roberge, M.; Geng, W.; Basler, J.; Chin, R.; Liu, A.; Ueda, R.; Hodges, D.; Escandon, E.; et al. Characterization of a CC49-based single-chain fragment-β-lactamase fusion protein for antibody-directed enzyme prodrug therapy (ADEPT). Bioconjug. Chem 2006, 17, 410–418. [Google Scholar]
- Kreitman, R.J.; Tallman, M.S.; Robak, T.; Coutre, S.; Wilson, W.H.; Stetler-Stevenson, M.; Fitzgerald, D.J.; Lechleider, R.; Pastan, I. Phase I trial of anti-CD22 recombinant immunotoxin moxetumomab pasudotox (CAT-8015 or HA22) in patients with hairy cell leukemia. J. Clin. Oncol 2012. [Google Scholar] [CrossRef]

| Name | Target | Format | Phase | Indication |
|---|---|---|---|---|
| 1D09C3 | HLA-DR | H IgG4 | I | Hodgkin’s lymphoma, myeloma |
| Lexatumumab | TRAIL-R2 | H IgG1 | I | Advanced solid tumors |
| Moxetumomab pasudotox | CD22 | Recombinant immunotoxin | I,II | Acute lymphoblastic leukemia, hairy cell leukemia, non-Hodgkin lymphoma |
| Fresolimumab | TGFβ | H IgG4 | I,II | Breast cancer; gliomas, kidney cancer; melanoma; mesothelioma. |
| Cixutumumab | IGF1R | H IgG1 | I,II | Adrenocortical carcinoma; breast cancer; colorectal cancer; HNC; islet cell cancer; liver cancer; Malignant Fibrous Histiocytoma neuroendocrine cancer; NSCLC; pancreatic cancer; prostate cancer; sarcoma; SCLC; solid tumors. |
| Mapatumumab | TRAIL-R1 | H IgG1 | II | Advanced Cervical Cancer, Hepatocellular Carcinoma, Multiple Myeloma |
| Necitumumab | EGFR | H IgG1 | III | NSCLC |
| Ramucirumab | VEGFR2 | H IgG1 | III | Hepatocellular carcinoma, metastatic gastric or gastresophageal junction adenocarcinoma |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dantas-Barbosa, C.; De Macedo Brigido, M.; Maranhao, A.Q. Antibody Phage Display Libraries: Contributions to Oncology. Int. J. Mol. Sci. 2012, 13, 5420-5440. https://doi.org/10.3390/ijms13055420
Dantas-Barbosa C, De Macedo Brigido M, Maranhao AQ. Antibody Phage Display Libraries: Contributions to Oncology. International Journal of Molecular Sciences. 2012; 13(5):5420-5440. https://doi.org/10.3390/ijms13055420
Chicago/Turabian StyleDantas-Barbosa, Carmela, Marcelo De Macedo Brigido, and Andrea Queiroz Maranhao. 2012. "Antibody Phage Display Libraries: Contributions to Oncology" International Journal of Molecular Sciences 13, no. 5: 5420-5440. https://doi.org/10.3390/ijms13055420