Glutamine Synthetase in Legumes: Recent Advances in Enzyme Structure and Functional Genomics
Abstract
:1. Introduction
1.1. Nitrogen Assimilation and Remobilization in Legumes
1.2. Glutamine Synthetase and Related Enzymes
2. Advances in Glutamine Synthetase Research
2.1. Enzyme Structure
2.2. Use of Mutants for the Study of Plastidic GS Functionality
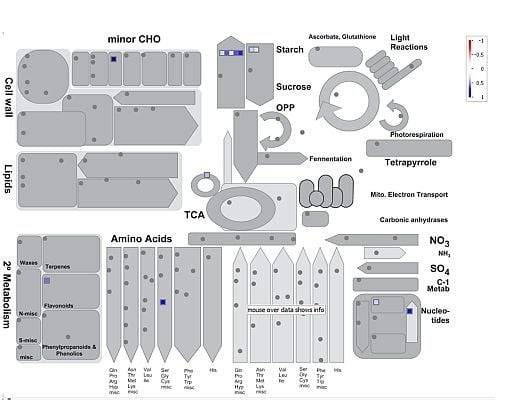
2.2.1. Plastidic GS and Photorespiration Transcriptomics
2.2.2. Plastidic GS and Nitrogen Nutrition
2.2.3. Plastidic GS, Photorespiration and Nodulation
2.2.4. Plastidic GS and Drought Stress Transcriptomics
2.2.5. Co-Expression Analysis of L. japonicus Plastidic GS
2.2.6. Analysis of Heterozygous Plastid GS Mutant Plants
3. Conclusions
Acknowledgements
References
- Good, A.G.; Shrawat, A.K.; Muench, D.G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 2004, 9, 597–605. [Google Scholar]
- Hirel, B.; Le Gouis, J.; Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot 2007, 58, 2369–2387. [Google Scholar]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot 2010, 105, 1141–1157. [Google Scholar]
- Keys, A.J.; Bird, I.F.; Cornelius, M.J.; Lea, P.J.; Wallsgrove, R.M.; Miflin, B.J. The photorespiratory nitrogen cycle. Nature 1978, 275, 741–743. [Google Scholar]
- Scott, D.B.; Farnden, K.J.F.; Robertson, J.G. Ammonia assimilation in lupin nodules. Nature 1976, 263, 703–705. [Google Scholar]
- Andrews, M. Nitrate and reduced-N concentrations in the xylem sap of Stellaria media, Xanthium strumarium and six legume species. Plant Cell Environ 1986, 9, 605–608. [Google Scholar]
- Do Amarante, L.; Lima, L.D.; Sodek, L. Growth and stress conditions cause similar changes in xylem amino acids for different legume species. Environ. Exp. Bot 2006, 58, 123–129. [Google Scholar]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol 2003, 131, 872–877. [Google Scholar]
- Udvardi, M.K.; Tabata, S.; Parniske, M.; Stougaard, J. Lotus japonicus: Legume research in the fast lane. Trends Plant Sci 2005, 10, 222–228. [Google Scholar]
- Márquez, A.J. (Ed.) Lotus Japonicus Handbook, 1st ed; Springer: Dordrecht, The Netherlands, 2005; pp. 1–384.
- Mathesius, U.; Journet, E.P.; Sumner, L.W. (Eds.) The Medicago Truncatula Handbook. Available online: http://www.noble.org/MedicagoHandbook/ accessed on 28 March 2012.
- Stacey, G.; Libault, M.; Brechenmacher, L.; Wan, J.; May, G.D. Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol 2006, 9, 110–121. [Google Scholar]
- Márquez, A.J.; Orea, A.; Pajuelo, P.; Pajuelo, E.; Romero, J.M.; Arcondéguy, T.; Betti, M.; García-Calderón, M.; Estivill, G.; Pal’ove-Balang, P. Nitrogen assimilation in roots of the model legume Lotus japonicus. Biologia 2004, 59, S69–S76. [Google Scholar]
- Márquez, A.J.; Betti, M.; García-Calderón, M.; Pal’ove-Balang, P.; Díaz, P.; Monza, J. Nitrate assimilation in Lotus japonicus. J. Exp. Bot 2005, 56, 1741–1749. [Google Scholar]
- Forde, B.G.; Cullimore, J.V. The molecular biology of glutamine synthetase in higher plants. Oxford Surv. Plant Mol. Cell. Biol 1989, 6, 247–296. [Google Scholar]
- Lam, H.G.; Coschigano, K.T.; Oliveira, I.C.; Melo-Oliveira, R.; Coruzzi, G.M. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol 1996, 47, 569–593. [Google Scholar]
- Cren, M.; Hirel, B. Glutamine synthetase in higher plants: Regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol 1999, 40, 1187–1193. [Google Scholar]
- Lea, P.J.; Ireland, P.J. Nitrogen Metabolism in Higher Plants. In Plant Amino Acids: Biochemistry and Biotechnology; Singh, B.K., Ed.; Marcel Dekker Inc: New York, NY, USA, 1999; pp. 1–47. [Google Scholar]
- Cánovas, F.M.; Ávila, C.; Cantón, F.R.; Cañas, R.A.; de la Torre, F. Ammonium assimilation and amino acid metabolism in conifers. J. Exp. Bot 2007, 58, 2307–2318. [Google Scholar]
- Bernard, S.M.; Habash, D.Z. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol 2009, 182, 608–620. [Google Scholar]
- Teixeira, J.; Pereira, S.; Cánovas, F.; Salema, R. Glutamine synthetase of potato (Solanum tuberosum L. cv. desiree) plants: Cell- and organ-specific expression and differential developmental regulation reveal specific roles in nitrogen assimilation and mobilization. J. Exp. Bot 2005, 56, 663–671. [Google Scholar]
- Tabuchi, M.; Abiko, T.; Yamaya, T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot 2007, 58, 2319–2327. [Google Scholar]
- Wallsgrove, R.M.; Turner, J.C.; Hall, N.P.; Kendall, A.C.; Bright, S.W. Barley mutants lacking chloroplast glutamine synthetase. Biochemical and genetic analysis. Plant Physiol 1987, 83, 155–158. [Google Scholar]
- Orea, A.; Pajuelo, P.; Pajuelo, E.; Quidiello, C.; Romero, J.M.; Márquez, A.J. Isolation of photorespiratory mutants from Lotus japonicus deficient in glutamine synthetase. Physiol. Plant 2002, 115, 352–361. [Google Scholar]
- Woodall, J.; Forde, B.G. Glutamine synthetase polypeptides in the roots of 55 legume species in relation to their climatic origin and the partitioning of nitrate assimilation. Plant Cell Environ 1996, 19, 848–858. [Google Scholar]
- Melo, P.M.; Lima, L.M.; Santos, I.M.; Carvalho, H.G.; Cullimore, J.V. Expression of the plastid-located glutamine synthetase of Medicago truncatula. Accumulation of the precursor in root nodules reveals an in vivo control at the level of protein import into plastids. Plant Physiol 2003, 132, 390–399. [Google Scholar]
- Taira, M.; Valtersson, U.; Burkhardt, B.; Ludwig, R.A. Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 2004, 16, 2048–2058. [Google Scholar]
- Betti, M.; Arcondéguy, T.; Márquez, A.J. Molecular analysis of two mutants from Lotus japonicus deficient in plastidic glutamine synthetase: Functional properties of purified GLN2 enzymes. Planta 2006, 224, 1068–1079. [Google Scholar]
- Sandal, N.; Petersen, T.R.; Murray, J.; Umehara, Y.; Karas, B.; Yano, K.; Kumagai, H.; Yoshikawa, M.; Saito, K.; Hayashi, M.; et al. Genetics of symbiosis in L. japonicus: Recombinant inbred lines, comparative genetic maps and map position on 35 symbiotic loci. Mol. Plant Microbe Int 2006, 19, 80–91. [Google Scholar]
- Seabra, A.R.; Vieira, C.P.; Cullimore, J.V.; Carvalho, H.G. Medicago truncatula contains a second gene encoding a plastid located glutamine synthetase exclusively expressed in developing seeds. BMC Plant Biol 2010, 10. [Google Scholar] [CrossRef]
- Vanoni, M.; Dossena, L.; van den Heuvel, R.; Curti, B. Structure-function studies of on the complex iron-sulfur flavoprotein glutamate synthase: The key enzyme of ammonia assimilation. Photosynth. Res 2005, 83, 219–238. [Google Scholar]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation and signalling. J. Exp. Bot 2007, 58, 2339–2358. [Google Scholar]
- Almassy, R.J.; Janson, C.A.; Hamlin, R.; Xuong, N.H.; Eisenberg, D. Novel subunit-subunit interactions in the structure of glutamine synthetase. Nature 1986, 323, 304–309. [Google Scholar]
- Yamashita, M.M.; Almassy, R.; Janson, C.; Causcio, D.; Eisenberg, D. Refined atomic model of glutamine synthetase at 3.5 Å resolution. J. Biol. Chem 1989, 264, 17681–17690. [Google Scholar]
- Gill, H.S.; Eisenberg, D. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry 2001, 40, 1903–1912. [Google Scholar]
- Gill, H.S.; Pfluegl, G.M.U.; Eisenberg, D. Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation. Biochemistry 2002, 41, 9863–9872. [Google Scholar]
- McParland, R.H.; Guevara, J.G.; Becker, R.R.; Evans, H.J. The purification and properties of the glutamine synthetase from the cytosol of soya-bean root nodules. Biochem. J 1976, 153, 597–606. [Google Scholar]
- Pushkin, A.V.; Tsuprun, V.L.; Dzhokharidze, T.Z.; Evstigneeva, Z.G.; Kretovich, W.L. Glutamine synthetase from the pumpkin leaf cytosol. Biochim. Biophys. Acta 1981, 662, 160–162. [Google Scholar]
- Pushkin, A.V.; Antoniuk, L.P.; Solovieva, N.A.; Shubin, V.V.; Evstigneeva, Z.G.; Kretovich, W.L.; Cherednikova, T.V.; Tsuprun, V.L.; Zograf, O.N.; Kiselev, N.A. Glutamine synthetase of lea leaf and seed cytosol. Structure and properties. Biochim. Biophys. Acta 1985, 828, 336–350. [Google Scholar]
- Tsuprun, V.L.; Zograf, O.N.; Orlova, E.V.; Kiselev, N.A.; Pushkin, A.V.; Shiffelova, G.E.; Solovieva, N.A.; Evstigneeva, Z.G.; Kretovich, W.C. Electron microscopy of multiple isoforms of glutamine synthetase from bacteroids and the cytosol of yellow lupin root nodules. Biochim. Biophys. Acta 1987, 913, 368–376. [Google Scholar]
- Boksha, I.S.; Schonfeld, H.J.; Langen, H.; Muller, F.; Tereshkina, E.B.; Burbaeva, G.S. Glutamine synthetase isolated from human brain: Octameric structure and homology of partial primary structure with human livel glutamine synthetase. Biochem. Mosc 2002, 67, 1012–1020. [Google Scholar]
- Llorca, O.; Betti, M.; González, J.M.; Valencia, A.; Márquez, A.J.; Valpuesta, J.M. The three-dimensional structure of an eukaryotic glutamine synthetase: Functional implications of its oligomeric structure. J. Struct. Biol 2006, 156, 469–479. [Google Scholar]
- Unno, H.; Uchida, T.; Sugawara, H.; Kurisu, G.; Sugiyama, T.; Yamaya, T.; Sakakibara, H.; Hase, T.; Kusunoki, M. Atomic structure of plant glutamine synthetase: A key enzyme for plant productivity. J. Biol. Chem 2006, 281, 29287–29296. [Google Scholar]
- Krajewski, W.W.; Collins, R.; Holmberg-Schiavone, L.; Jones, T.A.; Karlberg, T.; Mowbray, S.L. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J. Mol. Biol 2008, 375, 217–228. [Google Scholar]
- He, Y.; Gui, L.; Liu, Y.; Du, Y.; Zhou, Y.; Li, P.; Zhou, C. Crystal structure of Saccharomyces cerevisiae glutamine synthetase Gln1 suggests a nanotube-like supramolecular assembly. Proteins 2009, 76, 249–254. [Google Scholar]
- Seabra, A.R.; Carvalho, H.; Pereira, P.J. Crystallization and preliminary crystallographic characterization of glutamine synthetase from Medicago truncatula. Acta Crustallogr. F 2009, 65, 1309–1312. [Google Scholar]
- Estivill, G.; Guardado, P.; Buser, R.; Betti, M.; Márquez, A.J. Identification of an essential cysteinyl residue for the quaternary structure of glutamine synthetase α from Phaseolus vulgaris. Planta 2010, 231, 1101–1111. [Google Scholar]
- Estivill, G. Análisis Mutacional de la Estructura-función de las Glutamina Sintetasas de Plantas. Ph.D. Dissertation, University of Seville, Seville, Spain, 2011. [Google Scholar]
- Clemente, M.T.; Márquez, A.J. Functional importante of Asp56 from the α-polypeptide of Phaseolus vulgaris glutamine synthetase. An essential residue for transferase but not for bioysnthetic enzyme activity. Eur. J. Biochem 1999, 264, 453–460. [Google Scholar]
- Clemente, M.T.; Márquez, A.J. Site-directed mutagenesis of Glu-297 from the α-polypeptide of Phaseolus vulgaris glutamine synthetase alters kinetic and structural properties and confers resistance to l-methionine sulfoximine. Plant Mol. Biol 1999, 40, 835–845. [Google Scholar]
- Clemente, M.T.; Márquez, A.J. Site-directed mutagenesis of Cys-92 from the α-polypeptide of Phaseolus vulgaris glutamine synthetase reveals that this highly conserved residue is not essential for enzyme activity but it is involved in thermal stability. Plant Sci 2000, 154, 189–197. [Google Scholar]
- Betti, M.; Márquez, A.J.; Yanes, C.; Maestre, A. ATP binding to purified homopolymeric plant glutamine synthetase studied by isothermal titration calorimetry. Thermochim. Acta 2002, 394, 63–71. [Google Scholar]
- Spoerri, L. Post-Translational Regulation of Glutamine Synthetase inLotus japonicus: A Role for Mg2+ and Quaternary Structure. M.Sc. Thesis, University of Seville, Seville, Spain, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 2007. [Google Scholar]
- Finnemann, J.; Schoerring, J.K. Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant J 2000, 24, 171–181. [Google Scholar]
- Riedel, J.; Tischner, R.; Mäck, G. The chloroplastic glutamine synthetase (GS-2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta 2001, 213, 396–401. [Google Scholar]
- Pozuelo, M.; MacKintosh, C.; Galván, A.; Fernández, E. Cytosolic glutamine synthetase and not nitrate reductase from the green alga Chlamydomonas reinhardtii is phosphorylated and binds 14-3-3 proteins. Planta 2001, 212, 264–269. [Google Scholar]
- Lima, L.M.; Seabra, A.; Melo, P.M.; Cullimore, J.V.; Carvalho, H.G. Phosphorylation and subsequent interaction with 14-3-3 proteins regulate plastid glutamine synthetase in Medicago truncatula. Planta 2006, 223, 558–567. [Google Scholar]
- Lima, L.M.; Seabra, A.; Melo, P.M.; Cullimore, J.V.; Carvalho, H.G. Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. J. Exp. Bot 2006, 57, 2751–2761. [Google Scholar]
- Elrouby, N.; Coupland, G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA 2010, 107, 17415–17420. [Google Scholar]
- Melo, P.M.; Silva, L.S.; Ribeiro, I.; Seabra, A.R.; Carvalho, H.G. Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol 2011, 157, 1505–1517. [Google Scholar]
- Limami, A.R.; Phillipson, B.; Ameziane, R.; Pernollet, N.; Juang, Q.; Roy, R.; Deleens, E.; Chaumont-Bonnet, M.; Greshoff, P.M.; Hirel, B. Does root glutamine synthetase control plant biomass production in Lotus japonicus L.? Planta 1999, 209, 495–502. [Google Scholar]
- Ortega, J.L.; Temple, S.J.; Sengupta-Gopalan, C. Constitutive overexpression of cytosolic glutamine synthetase (GS1) gene in transgenic alfalfa demonstrates that GS1 may be regulated at the level of RNA stability and protein turnover. Plant Physiol 2001, 126, 109–121. [Google Scholar]
- Fei, H.; Chaillou, S.; Hirel, B.; Mahon, J.D.; Vessey, J.K. Overexpression of a soybean cytosolic glutamine synthetase gene linked to organ-specific promoters in pea plants grown in different concentrations of nitrate. Planta 2003, 216, 467–474. [Google Scholar]
- Ortega, J.L.; Moguel-Esponda, S.; Potenza, C.; Conclin, C.F.; Quintana, A.; Sengupta-Gopalan, C. The 3′ untranslated region of a soybean cytosolic glutamine synthetase (GS1) affects transcript stability and protein accumulation in transgenic alfalfa. Plant J 2006, 45, 832–846. [Google Scholar]
- Blackwell, R.D.; Murray, A.J.S.; Lea, P.J. Inhibition of photosynthesis in barley with decreased levels of chloroplastic glutamine synthetase activity. J. Exp. Bot 1987, 196, 1799–1809. [Google Scholar]
- Kozaki, A.; Takeba, G. Photorespiration protects C3 plants from photooxidation. Nature 1996, 384, 557–560. [Google Scholar]
- Somerville, C.R.; Ogren, W.L. A phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature 1979, 280, 833–836. [Google Scholar]
- Somerville, C.R.; Ogren, W.L. Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature 1980, 286, 257–59. [Google Scholar]
- Kendall, A.C.; Wallsgrove, R.M.; Hall, N.P.; Turner, J.C.; Lea, P.J. Carbon and nitrogen metabolism in barley (Hordeum vulgare L.) mutants lacking ferredoxin-dependent glutamate synthase. Planta 1986, 168, 316–323. [Google Scholar]
- Ferrario-Méry, S.; Hodges, M.; Hirel, B.; Foyer, C.H. Photorespiration-dependent increases in phosphoenolpyruvate carboxylase, isocitrate dehydrogenase and glutamate dehydrogenase in transformed tobacco plants deficient in ferredoxin-dependent glutamine-α-ketoglutarate aminotransferase. Planta 2002, 214, 877–886. [Google Scholar]
- Wingler, A.; Lea, P.J.; Quick, P.; Leegood, R.C. Photorespiration: Metabolic pathways and their role in stress protection. Philos. T. Roy. Soc. B 2000, 355, 1517–1529. [Google Scholar]
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, partners and origin. Trends Plant Sci 2010, 15, 330–336. [Google Scholar]
- Foyer, C.H.; Bloom, A.; Queval, G.; Noctor, G. Photorespiratory Metabolism: Genes, Mutants, Energetics and Redox Signalling. Annu. Rev. Plant Biol 2009, 60, 455–484. [Google Scholar]
- Hernández, I.; Alegre, L.; van Breusegem, F.; Munné-Bosch, S. How relevant are flavonoids as antioxidants in plants. Trends Plant Sci 2008, 14, 125–132. [Google Scholar]
- De León, I.P.; Sanz, A.; Hamberg, M.; Castresana, C. Involvement of the Arabidopsis α-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J 2002, 29, 61–72. [Google Scholar]
- Keys, A.J. The re-assimilation of ammonia produced by photorespiration and the nitrogen economy of C3 plants. Photosynth. Res 2006, 87, 165–175. [Google Scholar]
- Britto, D.T.; Kronzucker, H.J. NH4+ toxicity in higher plants: A critical review. J. Plant Physiol 2002, 159, 567–584. [Google Scholar]
- Lopes, M.; Araus, J.L. Comparative genomic and physiological analysis of nutrient response to NH4+, NH4+:NO3− and NO3− in barley seedlings. Physiol. Plant 2008, 134, 134–150. [Google Scholar]
- Migge, A.; Carrayol, E.; Hirel, B.; Becker, T.W. Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 2000, 210, 252–260. [Google Scholar]
- Chanh Ta, T.; Joy, K.W.; Ireland, R.J. Role of asparagine in the photorespiratory nitrogen metabolism of pea leaves. Plant Physiol 1985, 78, 334–337. [Google Scholar]
- García-Calderón, M.; Chiurazzi, M.; Espuny, R.; Márquez, A.J. Photorespiratory metabolism and nodule function: Behaviour of Lotus japonicus mutants deficient in plastid glutamine synthetase. Mol. Plant Microbe 2012, 25, 211–219. [Google Scholar]
- Harrison, J.; Pou de Crescenzo, M.A.; Sené, O.; Hirel, B. Does lowering glutamine synthetase activity in nodules modify nitrogen metabolism and growth of Lotus japonicus? Plant Physiol 2003, 133, 253–262. [Google Scholar]
- Ortega, J.L.; Temple, S.J.; Bagga, S.; Ghoshroy, S.; Sengupta-Gopalan, C. Biochemical and molecular characterization of transgenic Lotus japonicus plants constitutively over-expressing a cytosolic glutamine synthetase gene. Planta 2004, 219, 807–818. [Google Scholar]
- Carvalho, H.; Sunkel, C.; Salema, R.; Cullimore, J.V. Heteromeric assembly of the cytosolic glutamine synthetase polypeptides of Medicago truncatula: Complementation of a glnA Escherichia coli mutant with a plant domain-swapped enzyme. Plant Mol. Biol 1997, 35, 623–632. [Google Scholar]
- López, M.; Herrera-Cervera, J.A.; Iribarne, C.; Tejera, N.A.; Lluch, C. Growth and nitrogen fixation in Lotus japonicus and Medicago truncatula under NaCl stress: Nodule carbon metabolism. J. Plant Physiol 2008, 165, 641–650. [Google Scholar]
- Gordon, A.J.; Minchin, F.R.; James, C.L.; Komina, O. Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 1999, 120, 867–877. [Google Scholar]
- Phillips, D.A.; Newell, K.D.; Hassell, S.A.; Felling, C.E. The effect of CO2 enrichment on root nodule development and symbiotic N2 reduction in Pisum sativum L. Am. J. Bot 1976, 63, 356–362. [Google Scholar]
- Finn, G.A.; Brun, W.A. Effect of atmospheric CO2 enrichment on growth, non-structural carbohydrate content, and root nodule activity in soybean. Plant Physiol 1982, 69, 327–331. [Google Scholar]
- Ortega, J.L.; Sánchez, F.; Soberón, M.; Lara-Flores, M. Regulation of nodule glutamine synthetase by CO2 levels in bean (Phaseolus vulgaris L.). Plant Physiol 1992, 98, 584–587. [Google Scholar]
- Cabrerizo, P.M.; González, E.M.; Aparicio-Tejo, P.M.; Arrese-Igor, C. Continuous CO2 enrichment leads to increased nodule biomass, carbon availability to nodules and activity of carbon-metabolising enzymes but does not enhance specific nitrogen fixation in pea. Physiol. Plant 2001, 113, 33–40. [Google Scholar]
- Fischinger, S.A.; Hristozkova, M.; Mainassara, Z.-A.; Schulze, J. Elevated CO2 concentration around alfalfa nodules increases N2 fixation. J. Exp. Bot 2010, 61, 121–130. [Google Scholar]
- Vance, C.P.; Heichel, G.H. Carbon in N2 fixation: Limitation or exquisite adaptation. Annu. Rev. Plant Physiol 1991, 42, 373–392. [Google Scholar]
- Cordoba, E.; Shishkova, S.; Vance, C.; Hernández, G. Antisense inhibition of NADH-glutamate synthase impairs carbon/nitrogen assimilation in nodules of alfalfa. Plant J 2003, 33, 1037–1049. [Google Scholar]
- Kinoshita, H.; Nagasaki, J.; Yoshikawa, N.; Yamamoto, A.; Takito, S.; Kawasaki, M.; Sugiyama, T.; Miyake, H.; Weber, A.P.M.; Taniguchi, M. The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J 2011, 65, 15–26. [Google Scholar]
- Omrane, S.; Ferrarini, A.; D’Apuzzo, E.; Rogato, A.; Delledonne, M.; Chiurazzi, M. Symbiotic competence in Lotus japonicus is affected by plant nitrogen status: Transcriptomic identification of genes affected by a new signalling pathway. New Phytol 2009, 183, 380–394. [Google Scholar]
- Omrane, S.; Chiurazzi, M. A variety of regulatory mechanisms are involved in the nitrogen-dependent modulation of the nodule organogenesis program in legume roots. Plant Signal. Behav 2009, 4, 1066–1068. [Google Scholar]
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci 2005, 24, 23–58. [Google Scholar]
- Brugiére, N.; Dubois, F.; Limami, A.M.; Lelandais, M.; Roux, Y.; Sangwan, R.S.; Hirel, B. Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 1999, 11, 1995–2012. [Google Scholar]
- Hoshida, H.; Tanaka, Y.; Hibino, T.; Hayashi, Y.; Tanaka, A.; Takabe, T.; Takabe, T. Enhanced tolerance to salt stress in transgenic rice that overexpress chloroplast glutamine synthetase. Plant Mol. Biol 2000, 43, 103–111. [Google Scholar]
- Díaz, P.; Betti, M.; Sánchez, D.H.; Udvardi, M.K.; Monza, J.; Márquez, A.J. Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol 2010, 188, 1001–1013. [Google Scholar]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci 2010, 15, 89–97. [Google Scholar]
- Acharya, B.R.; Assmann, S.M. Hormone interactions in stomatal function. Plant Mol. Biol 2009, 69, 451–462. [Google Scholar]
- Bouché, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci 2004, 9, 110–115. [Google Scholar]
- Hiroyuki, K.; Terauchi, R. Regulation of expression of rice thaumatin-like protein: Inducibility by elicitor requires promoter W-box elements. Plant Cell Rep 2008, 27, 1521–1528. [Google Scholar]
- Guo, Y.; Huang, C.; Xie, Y.; Song, F.; Zhou, X. A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stress. Planta 2010, 232, 1499–1509. [Google Scholar]
- Perry, J.A.; Wang, T.L.; Welham, T.J.; Gardner, S.; Pike, J.M.; Yoshida, S.; Parniske, M. A TILLING reverse genetic tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol 2003, 131, 866–871. [Google Scholar]
- Urbanski, D.F.; Małolepszy, A.; Stougaard, J.; Andersen, S.U. Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J 2012, 69, 731–741. [Google Scholar]
- Usadel, B.; Obayashi, T.; Mutwil, M.; Giorgi, F.M.; Bassel, G.W.; Tanimoto, M.; Chow, A.; Steinhouser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ 2009, 32, 1633–1651. [Google Scholar]
- Swarbreck, S.M.; Defoin-Platel, M.; Hindle, M.; Saqi, M.; Habash, D.Z. New perspectives on glutamine synthetase in grasses. J. Exp. Bot 2011, 62, 1511–1522. [Google Scholar]
- Høgslund, N.; Radutoiu, S.; Krusell, L.; Voroshilova, V.; Hannah, M.A.; Goffard, N.; Sánchez, D.H.; Lippold, F.; Ott, T.; Sato, S.; et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS One 2009, 4. [Google Scholar] [CrossRef]
- Goffard, N.; Weiller, G. Genebins: A database for classifying gene expression data, with application to plant genome arrays. BMC Bioinforma 2007, 8. [Google Scholar] [CrossRef]
- Herskowitz, I. Functional inactivation of genes by negative dominant mutations. Nature 1987, 329, 219–222. [Google Scholar]




| WT only | Shared | Ljgln2-2 Only | |
|---|---|---|---|
| Total Probesets | 655 | 825 | 5785 |
| Up-Regulated | 155 | 384 | 2470 |
| Down-Regulated | 500 | 441 | 3315 |
| WT | |||
|---|---|---|---|
| Probeset | log2 FC | Description | Similar to |
| Up-Regulated | |||
| chr2.TM0641.8 | 1.98 | NAC domain protein | AT1G69490 |
| chr5.CM0341.27 | 1.96 | AP2-EREBP Transcription factor | AT3G23240 |
| chr2.CM0191.49.2 | 1.91 | Polyketide reductase | AT1G59960 |
| Ljwgs_093636.1 | 1.87 | Alpha-dioxygenase | AT3G01420 |
| Ljwgs_050995.1 | 1.77 | Methyltransferase protein | AT3G11480 |
| Ljwgs_080010.1 | 1.73 | Pleiotropic drug resistance protein | AT1G15520 |
| chr4.CM0256.39 | 1.73 | Cytochrome P450 | AT4G37370 |
| chr2.CM0249.88 | 1.72 | Isoflavone reductase | AT4G39230 |
| TM0802.13 | 1.69 | 2-Hydroxyisoflavanone synthase | AT5G06900 |
| Ljwgs_018999.1 | 1.64 | Glutathione S-transferase | AT2G29420 |
| Down-regulated | |||
| Ljwgs_108871.1 | −2.74 | Hypothetical protein | AT3G02550 |
| Ljwgs_035693.2 | −2.46 | Hypothetical protein | AT3G20810 |
| Ljwgs_089359.1.1 | −2.02 | Early flowering 4 protein | AT2G40080 |
| chr3.CM0711.3.2 | −1.93 | Unknown protein | AT4G10270 |
| chr2.CM0191.60 | −1.83 | Nlj21 | - |
| Ljwgs_080939.1 | −1.81 | Beta-glucosidase like protein | AT2G44480 |
| chr3.TM0426.3 | −1.75 | Hypothetical protein | AT5G22580 |
| chr1.CM0398.23.1 | −1.66 | Gibberellin induced protein | AT1G74670 |
| chr3.CM0155.27 | −1.64 | Peroxidase | AT1G05260 |
| TC17223 | −1.64 | Hypothetical protein | - |
| WT | |||
| Ljgln2-2 | |||
| Probeset | log2 FC | Description | Similar to |
| Up-Regulated | |||
| Ljwgs_044797.1 | 5.37 | 60S ribosomal protein | AT1G26910 |
| chr3.CM0590.56 | 4.52 | Chalcone synthase | AT5G13930 |
| Ljwgs_036303.1 | 4.43 | NAC domain protein | AT4G27410 |
| TM0802.13 | 4.35 | 2-Hydroxyisoflavanone synthase | AT5G06900 |
| chr2.CM0250.2 | 4.34 | MYB transcription factor | AT4G37260 |
| gi45637799 | 4.26 | Hypothetical protein | - |
| chr2.CM0018.54 | 4.19 | Chalcone synthase | AT5G13930 |
| Ljwgs_099009.1 | 4.06 | Chalcone synthase | AT5G13930 |
| Ljwgs_093636.1 | 4.04 | Alpha-dioxygenase | AT3G01420 |
| chr5.CM0909.51 | 4.03 | Glutathione S-transferase | AT2G29420 |
| Down-regulated | |||
| chr1.CM0001.63 | −4.66 | Probable 2-Isopropylmalate synthase | AT1G74040 |
| Ljwgs_091497.1 | −2.97 | Myo-inositol-1-phosphate synthase | AT5G10170 |
| TM0810.14 | −2.88 | Cytochrome P450 | AT2G45550 |
| chr1.CM0398.23.1 | −2.80 | Gibberellin regulated protein | AT1G74670 |
| TM1614.14.1 | −2.60 | Hypothetical protein | AT1G59960 |
| Ljwgs_028558.1 | −2.55 | Pectate lyase | AT4G24780 |
| Ljwgs_062989.1 | −2.54 | Terpene synthase | AT4G16730 |
| chr3.CM0142.55 | −2.52 | Hypothetical protein | AT5G20190 |
| chr1.CM0001.70.2 | −2.45 | Hypothetical protein | AT5G13750 |
| Ljwgs_043433.1 | −2.35 | Benzoyl transferase | AT5G17540 |
| Name | Probeset | Nitrogen Source | ||
|---|---|---|---|---|
| NO3− | NH4+ | NH4NO3 | ||
| LjGS1.1 | TM0053.11 | 2.71 | 2.69 | 0.93 |
| LjGS1.2 | gi1246767 | 0.95 | 0.84 | 1.88 |
| LjGS1.3 | Ljwgs_019428.1 | 6.22 | 3.94 | 0.22 |
| LjGLN2 | gi18266052 | 8.99 | 4.96 | 0.19 |
| LjGLU1 | chr1.CM0009.24 | 4.77 | 2.64 | 0.25 |
| LjGLT1 | Ljwgs_035611.1 | 0.71 | 1.40 | 1.36 |
| LjGLT2 | Ljwgs_037992.1 | 1.06 | 1.70 | 1.23 |
| LjAS1 | gi897770 | 4.91 | 2.58 | 0.30 |
| LjAS2 | gi897772 | 4.57 | 3.64 | 0.73 |
| LjGDH3 | Ljwgs_035272.1 | 2.32 | 1.73 | 1.46 |
| LjGDH4 | Ljwgs_009442.1 | 1.09 | 1.04 | 1.13 |
| WT Only | Shared | Ljgln2-2 Only | |
|---|---|---|---|
| Total Probesets | 538 | 2070 | 5845 |
| Up-Regulated | 207 | 946 | 3636 |
| Down-Regulated | 331 | 1124 | 2209 |
| WT | |||
|---|---|---|---|
| Probeset | log2 FC | Description | Similar to |
| Up-Regulated | |||
| TC11101 | 4.83 | Glutamate decarboxylase | AT5G17330 |
| chr1.TM1635.18 | 4.77 | ACC synthase | AT3G61510 |
| Ljwgs_047159.1 | 4.61 | STIG1-related protein | AT1G11925 |
| chr5.CM0909.59 | 4.47 | Glutathione S-transferase | AT2G29420 |
| chr1.CM0141.2 | 4.37 | Nitrate/peptide transporter | AT1G32450 |
| Ljwgs_036708.1 | 4.32 | Pectinesterase | AT2G45220 |
| chr4.CM0429.5 | 4.32 | Mitochondrial inner membr. translocase | AT4G16160 |
| chr5.CM0089.120 | 4.15 | Inositol-1,4,5-trisphosphate 5-phosphatase | AT1G47510 |
| chr5.CM0148.50.2 | 4.11 | Cytochrome P450 | AT5G52400 |
| TM0763.11 | 4.08 | 12-oxophytodienoate reductase | AT2G06050 |
| Down-regulated | |||
| Ljwgs_040576.1 | −4.15 | Glutaredoxin | AT5G18600 |
| Ljwgs_056053.1 | −4.15 | Alpha-expansin family protein | AT2G39700 |
| Ljwgs_006332.1 | −3.92 | Hypothetical protein | AT1G30260 |
| chr1.BM1732.18 | −3.82 | Hypothetical protein | AT3G11210 |
| BM0976.11 | −3.78 | Hypothetical protein | AT2G01050 |
| WT | |||
| Ljwgs_028040.1 | −3.74 | Ammonium transporter | AT4G13510 |
| chr1.CM0233.42 | −3.47 | Nucleic acid binding protein | AT1G52950 |
| chr6.TM1374.27 | −3.41 | hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase-like | AT2G19070 |
| Ljwgs_016759.2 | −3.41 | Chloride channel protein | AT5G40890 |
| TM1490.11 | −3.35 | MYB transcription factor | AT2G21650 |
| Ljgln2-2 | |||
| Probeset | log2 FC | Description | Similar to |
| Up-Regulated | |||
| chr1.CM0012.67 | 8.20 | Thaumatin-like protein | AT1G20030 |
| Ljwgs_075692.1.1 | 7.87 | GDSL esterase/lipase | AT1G29660 |
| chr3.TM1465.12 | 7.50 | SPX domain-containing protein | AT2G45130 |
| chr1.TM1635.18 | 7.34 | ACC synthase | AT3G61510 |
| Ljwgs_036708.1 | 6.93 | Pectinesterase | AT2G45220 |
| chr2.CM1150.57 | 6.84 | Metalloendoproteinase | AT1G70170 |
| chr1.CM0104.32 | 6.84 | NAC domain transcription factor | AT3G04070 |
| Ljwgs_020980.2 | 6.80 | C2H2 transcription factor | AT2G37430 |
| Ljwgs_047159.1 | 6.73 | STIG1-related protein | AT1G11925 |
| Ljwgs_145133.1 | 6.72 | Late embryogenesis abundant (LEA) | AT3G53040 |
| Down-regulated | |||
| chr1.BM1732.18 | −8.81 | Lipase/hydrolase protein | AT3G11210 |
| chr2.CM0249.113 | −7.27 | Myb familily transcription factor | AT2G19510 |
| Ljwgs_040576.1 | −6.77 | Glutaredoxin | AT5G18600 |
| Ljwgs_073999.0.1 | −6.70 | Cytochrome P450 | AT1G24180 |
| Ljwgs_052903.1 | −6.12 | Lipoxygenase | AT1G55020 |
| Ljwgs_058749.1 | −5.73 | Beta-glucosidase | AT5G42260 |
| Ljwgs_127990.1 | −5.62 | Myb-related transcription factor | AT4G39250 |
| TM0990.31.1 | −5.59 | Hypothetical protein | - |
| chr3.CM0590.43 | −5.55 | Hypothetical protein | AT4G27450 |
| Ljwgs_091781.1 | −5.45 | Lipoxygenase | AT1G55020 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Betti, M.; García-Calderón, M.; Pérez-Delgado, C.M.; Credali, A.; Estivill, G.; Galván, F.; Vega, J.M.; Márquez, A.J. Glutamine Synthetase in Legumes: Recent Advances in Enzyme Structure and Functional Genomics. Int. J. Mol. Sci. 2012, 13, 7994-8024. https://doi.org/10.3390/ijms13077994
Betti M, García-Calderón M, Pérez-Delgado CM, Credali A, Estivill G, Galván F, Vega JM, Márquez AJ. Glutamine Synthetase in Legumes: Recent Advances in Enzyme Structure and Functional Genomics. International Journal of Molecular Sciences. 2012; 13(7):7994-8024. https://doi.org/10.3390/ijms13077994
Chicago/Turabian StyleBetti, Marco, Margarita García-Calderón, Carmen M. Pérez-Delgado, Alfredo Credali, Guillermo Estivill, Francisco Galván, José M. Vega, and Antonio J. Márquez. 2012. "Glutamine Synthetase in Legumes: Recent Advances in Enzyme Structure and Functional Genomics" International Journal of Molecular Sciences 13, no. 7: 7994-8024. https://doi.org/10.3390/ijms13077994