Multiple Mechanisms and Challenges for the Application of Allopolyploidy in Plants
Abstract
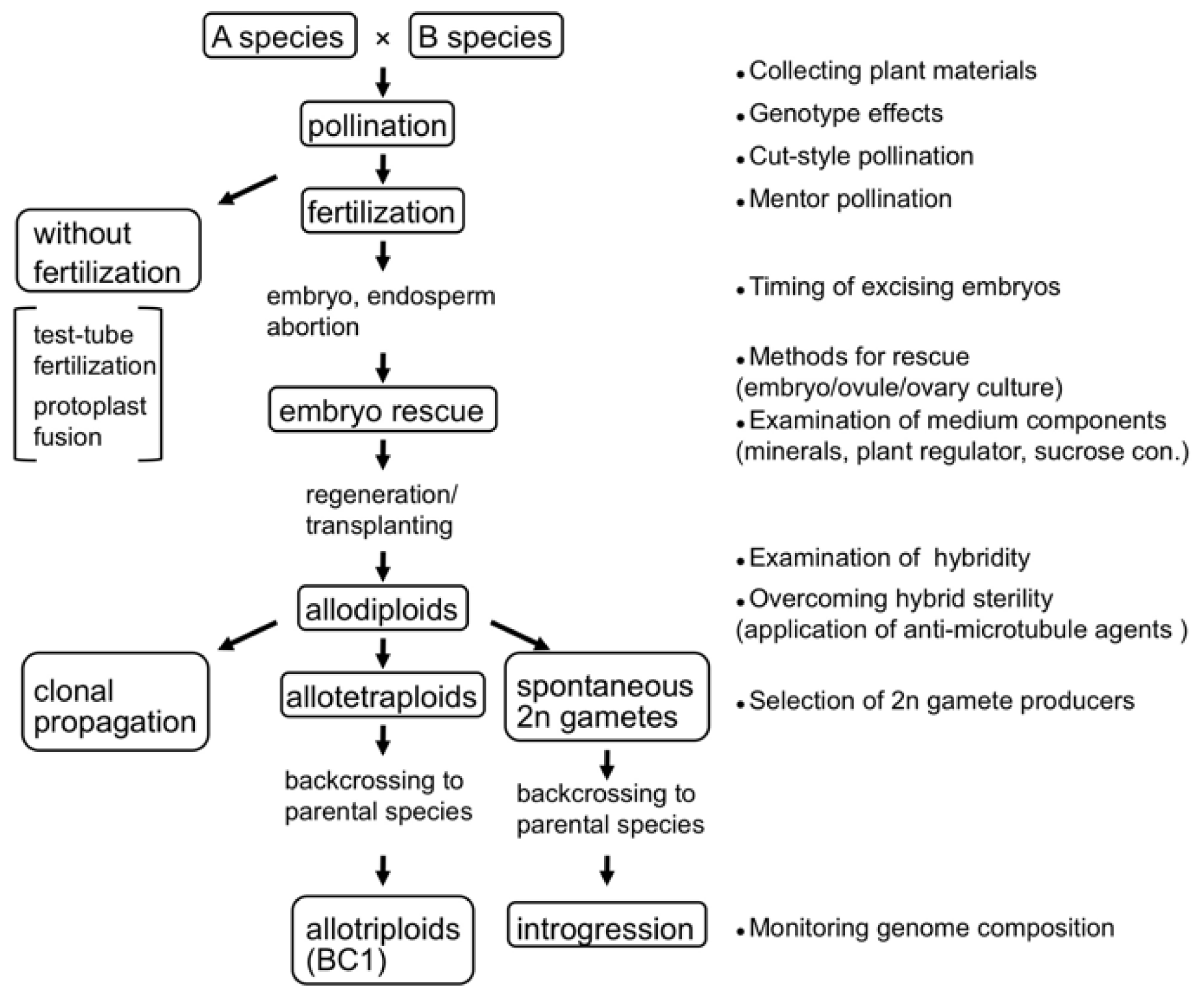
:1. Introduction
2. Techniques for Synthesizing Allopolyploids
3. Reproductive Barrier
3.1. Genomic Imprinting
3.2. Cytoplasmic Male Sterility
4. Self-Compatibility
5. Genetic and Epigenetic Changes
6. Changes in Homoeologous Gene Expression
7. Conclusion
Acknowledgments
References
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol 2005, 8, 135–141. [Google Scholar]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol 2007, 58, 377–406. [Google Scholar]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst 1998, 29, 467–501. [Google Scholar]
- Yamauchi, A.; Hosokawa, A.; Nagata, H.; Shimoda, M. Triploid bridge and role of parthenogenesis in the evolution of autopolyploidy. Am. Nat 2004, 164, 101–112. [Google Scholar]
- Soltis, D.E.; Soltis, P.S. Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci 1993, 12, 243–273. [Google Scholar]
- Griffiths, S.; Sharp, R.; Foote, T.N.; Bertin, I.; Wanous, M.; Reader, S.; Colas, I.; Moore, G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 2006, 439, 749–752. [Google Scholar]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet 2005, 6, 836–846. [Google Scholar]
- Osborn, T.C.; Pires, J.C.; Birchler, J.A.; Auger, D.L.; Chen, Z.J.; Lee, H.S.; Comai, L.; Madlung, A.; Doerge, R.W.; Colot, V.; et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet 2003, 19, 141–147. [Google Scholar]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 2010, 15, 57–71. [Google Scholar]
- Nasrallah, M.E.; Yogeeswaran, K.; Snyder, S.; Nasrallah, J.B. Arabidopsis species hybrids in the study of species differences and evolution of amphiploidy in plants. Plant Physiol 2000, 124, 1605–1614. [Google Scholar]
- Fujimoto, R.; Taylor, J.M.; Sasaki, T.; Kawanabe, T.; Dennis, E.S. Genome wide gene expression in artificially synthesized amphidiploids of Arabidopsis. Plant Mol. Biol 2011, 77, 419–431. [Google Scholar]
- Ni, Z.; Kim, E.D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2009, 457, 327–331. [Google Scholar]
- Guo, M.; Davis, D.; Birchler, J.A. Dosage effects on gene expression in a maize ploidy series. Genetics 1996, 142, 1349–1355. [Google Scholar]
- Ha, M.; Kim, E.D.; Chen, Z.J. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc. Natl. Acad. Sci. USA 2009, 106, 2295–2300. [Google Scholar]
- Okamoto, S.; Odashima, M.; Fujimoto, R.; Sato, Y.; Kitashiba, H.; Nishio, T. Self-compatibility in Brassica napus is caused by independent mutations in S-locus genes. Plant J 2007, 50, 391–400. [Google Scholar]
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar]
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol 2011, 52, 750–764. [Google Scholar]
- Wendel, J.F.; Cronn, R.C. Polyploidy and the evolutionary history of cotton. Adv. Agron 2003, 78, 139–186. [Google Scholar]
- Lee, J.J.; Woodward, A.W.; Chen, Z.J. Gene expression changes and early events in cotton fibre development. Ann. Bot 2007, 100, 1391–1401. [Google Scholar]
- Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot 1935, 7, 389–452. [Google Scholar]
- Kashkush, K.; Feldman, M.; Levy, A.A. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 2002, 160, 1651–1659. [Google Scholar]
- Gaeta, R.T.; Pires, J.C.; Iniguez-Luy, F.; Leon, E.; Osborn, T.C. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 2007, 19, 3403–3417. [Google Scholar]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic changes in synthetic Arabidopsis polyploids. Plant J 2005, 41, 221–230. [Google Scholar]
- Madlung, A.; Masuelli, R.W.; Watson, B.; Reynolds, S.H.; Davison, J.; Comai, L. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol 2002, 129, 733–746. [Google Scholar]
- Lukens, L.N.; Pires, J.C.; Leon, E.; Vogelzang, R.; Oslach, L.; Osborn, T. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol 2006, 140, 336–348. [Google Scholar]
- Beaulieu, J.; Jean, M.; Belzile, F. The allotetraploid Arabidopsis thaliana-Arabidopsis lyrata subsp. petraea as an alternative model system for the study of polyploidy in plants. Mol. Genet. Genomics 2009, 281, 421–435. [Google Scholar]
- Parisod, C.; Alix, K.; Just, J.; Petit, M.; Sarilar, V.; Mhiri, C.; Ainouche, M.; Chalhoub, B.; Glandbastien, M.A. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol 2010, 186, 37–45. [Google Scholar]
- Comai, L.; Tyagi, A.P.; Winter, K.; Holmes-Davis, R.; Reynolds, S.H.; Stevens, Y.; Byers, B. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 2000, 12, 1551–1568. [Google Scholar]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet 2003, 33, 102–106. [Google Scholar]
- He, P.; Friebe, B.R.; Gill, B.S.; Zhou, J.M. Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol. Biol 2003, 52, 401–414. [Google Scholar]
- Adams, K.L.; Percifield, R.; Wendel, J.F. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 2004, 168, 2217–2226. [Google Scholar]
- Albertin, W.; Balliau, T.; Brabant, P.; Chèvre, A.M.; Eber, F.; Malosse, C.; Thiellement, H. Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 2006, 173, 1101–1113. [Google Scholar]
- Wang, J.; Tian, L.; Lee, H.S.; Wei, N.E.; Jiang, H.; Watson, B.; Madlung, A.; Osborn, T.C.; Doerge, R.W.; Comai, L.; Chen, Z.J. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 2006, 172, 507–517. [Google Scholar]
- Gaeta, R.T.; Yoo, S.Y.; Pires, J.C.; Doerge, R.W.; Chen, Z.J.; Osborn, T.C. Analysis of gene expression in resynthesized Brassica napus allopolyploids using Arabidopsis 70mer oligo microarrays. PLoS One 2009, 4, e4760. [Google Scholar]
- Pumphrey, M.; Bai, J.; Laudencia-Chingcuanco, D.; Anderson, O.; Gill, B.S. Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics 2009, 181, 1147–1157. [Google Scholar]
- Rapp, R.A.; Udall, J.A.; Wendel, J.F. Genomic expression dominance in allopolyploids. BMC Biol 2009, 7, 18. [Google Scholar]
- Mejía-Jiménez, A.; Muñoz, C.; Jacobsen, H.J.; Roca, W.M.; Singh, S.P. Interspecific hybridization between common and tepary beans: Increased hybrid embryo growth, fertility, and efficiency of hybridization through recurrent and congruity backcrossing. Theor. Appl. Genet 1994, 88, 324–331. [Google Scholar]
- Akbar, M.A. Chromosomal stability and performance of resynthesized Brassica napus produced for gain in earliness and short-day response. Hereditas 1989, 111, 247–253. [Google Scholar]
- Van Eijk, J.P.; van Raamsdonk, L.W.D.; Eikelboom, W.; Bino, R.J. Interspecific crosses between Tulipa gesneriana cultivars and wild Tulipa species: A survey. Sex. Plant Reprod 1991, 4, 1–5. [Google Scholar]
- Murfett, J.; Strabala, T.J.; Zurek, D.M.; Mou, B.; Beecher, B.; McClure, B.A. S RNase and interspecific pollen rejection in the genus Nicotiana: Multiple pollen-rejection pathways contribute to unilateral incompatibility between self-Incompatible and self-compatible species. Plant Cell 1996, 8, 943–958. [Google Scholar]
- Asano, Y. Studies on crosses between distantly related species of lilies IV. The culture of immature hybrid embryos 0.3–0.4 mm long. J. Jpn. Soc. Hort. Sci 1980, 49, 114–118. [Google Scholar]
- Kho, Y.O.; Baër, J. Incompatibility problems in species crosses of tulips. Euphytica 1971, 20, 30–35. [Google Scholar]
- Franken, J.; Custers, J.B.M.; Bino, R.J. Effects of temperature on pollen tube growth and fruit set in reciprocal crosses between Cucumis sativus and C. metuliferus. Plant Breed 1988, 100, 150–153. [Google Scholar]
- Watts, V.M. Influence of intrastylar pollination on seed set in lilies. Proc. Am. Soc. Hort. Sci 1967, 91, 660–663. [Google Scholar]
- Asano, Y.; Myodo, H. Studies on crosses between distantly related species of lilies I. For the intrastylar pollination technique. J. Jpn. Soc. Hort. Sci 1977, 46, 59–65. [Google Scholar]
- Stettler, R.F. Irradiated mentor pollen: Its use in remote hybridization of black cottonwood. Nature 1968, 219, 746–747. [Google Scholar]
- Dayton, D.F. Overcoming self-incompatibility in plants with killed compatible pollen. J. Am. Soc. Hort. Sci 1974, 99, 190–192. [Google Scholar]
- Den Nijs, A.P.M.; Oost, E.H. Effects of mentor pollen on pollen-pistil incongruities among species of Cucumis L. Euphytica 1980, 29, 267–271. [Google Scholar]
- Visser, T. Pollen and pollination experiments IV. Mentor pollen and pioneer pollen techniques regarding incompatibility and incongruity in apple and pear. Euphytica 1981, 31, 305–312. [Google Scholar]
- Emsweller, S.L.; Stuart, N.W. Use of growth regulating substances to overcome incompatibilities in Lilium. Proc. Am. Soc. Hort. Sci 1948, 51, 581–589. [Google Scholar]
- Raghavan, V.; Srivastava, P.S. Embryo culture. In Experimental Embryology of Vascular Plants; Johri, B.M., Ed.; Springer-Verlag: Berlin, Germany, 1982; pp. 195–230. [Google Scholar]
- Williams, E.G.; Maheswaran, G.; Hutchinson, J.F. Embryo and ovule culture in crop improvement. Plant Breed. Rev 1987, 5, 181–236. [Google Scholar]
- Iwai, S.; Kishi, C.; Nakata, K.; Kawashima, N. Production of Nicotiana tabacum x Nicotiana acuminata hybrid by ovule culture. Plant Cell Rep 1986, 5, 403–404. [Google Scholar]
- Chen, B.Y.; Heneen, W.K.; Jonsson, R. Resynthesis of Brassica napus L. through interspecific hybridization between B. alboglabra Bailey and B. campestris L. with special emphasis on seed colour. Plant Breed 1986, 101, 52–59. [Google Scholar]
- Asano, Y.; Myodo, H. Studies on crosses between distantly related species of lilies II. The culture of immature hybrid embryos. J. Jpn. Soc. Hort. Sci 1977, 46, 267–273. [Google Scholar]
- Okazaki, K.; Asano, Y.; Oosawa, K. Interspecific hybrids between Lilium ‘Oriental’ hybrid and L. ‘Asiatic’ hybrid produced by embryo culture with revised media. Breed. Sci 1994, 44, 59–64. [Google Scholar]
- Gangadevi, T.; Rao, P.N.; Rao, B.H.; Satyanarayana, K.V. A study of morphology, cytology and sterility in interspecific hybrids and amphidiploids of Nicotiana knightiana × N. umbratica. Theor. Appl. Genet 1985, 70, 330–332. [Google Scholar]
- Poysa, V. The development of bridge lines for interspecific gene transfer between Lycopersicon esculentum and L. peruvianum. Theor. Appl. Genet 1990, 79, 187–192. [Google Scholar]
- Choudhary, B.R.; Joshi, P.; Ramarao, S. Interspecific hybridization between Brassica carinata and Brassica rapa. Plant Breed 2000, 119, 417–420. [Google Scholar]
- Asano, Y. Overcoming interspecific hybrid sterility in Lilium. J. Jpn. Soc. Hort. Sci 1982, 51, 75–81. [Google Scholar]
- Bretagnolle, F.; Thompson, J.D. Gametes with the somatic chromosome number: Mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol 1995, 129, 1–22. [Google Scholar]
- Lim, K.B.; Chung, J.D.; Kronenburg, B.C.E.; Ramanna, M.S.; de Jong, J.H.; Jacobsen, E.; van Tuyl, J.M. Introgression of Lilium rubellum Baker chromosomes into L. Longiflorum Thunb.: A genome painting study of the F1 hybrid, BC1 and BC2 progenies. Chromosome Res 2000, 8, 119–125. [Google Scholar]
- Nimura, M.; Kato, J.; Horaguchi, H.; Mii, M.; Sakai, K.; Katoh, T. Induction of fertile amphidiploids by artificial chromosome-doubling in interspecific hybrid between Dianthus caryophyllus L. and D. japonicus Thunb. Breed. Sci 2006, 56, 303–310. [Google Scholar]
- Jensen, C.J. Chromosome Doubling Techniques in Haploids. In Haploids in Higher Plants: Advances and Potential; Kasha, K.J., Ed.; University of Guelph: Guelph, ON, Canada, 1974; pp. 153–190. [Google Scholar]
- Van Tuyl, J.M.; Meijer, B.; van Diën, M.P. The use of oryzalin as an alternative for colchicine in in-vitro chromosome doubling of Lilium and Nerine. Acta Hort 1992, 352, 625–630. [Google Scholar]
- Kermani, M.J.; Sarasan, V.; Roberts, A.V.; Yokoya, K.; Wentworth, J.; Sieber, V.K. Oryzalin-induced chromosome doubling in Rosa and its effect on plant morphology and pollen fertility. Theor. Appl. Genet 2003, 107, 1195–1200. [Google Scholar]
- Burge, G.K.; Morgan, E.R.; Eason, J.R.; Clark, G.E.; Catley, J.L.; Seelye, J.F. Sandersonia aurantiaca: Domestication of a new ornamental crop. Sci. Hort 2008, 118, 87–99. [Google Scholar]
- Sree Ramulu, K.; Verhoeven, H.A.; Dijkhuis, P. Mitotic blocking, micronucleation, and chromosome doubling by oryzalin, amiprophos-methyl, and colchicine in potato. Protoplasma 1991, 160, 65–73. [Google Scholar]
- Dvorak, J.; Harvey, B.L.; Coulman, B.E. The use of nitrous oxide for producing euploids and aneuploids in wheat and barley. Can. J. Genet. Cytol 1973, 15, 205–214. [Google Scholar]
- Kato, A. Chromosome doubling of haploid maize seedling using nitrous oxide gas at the flower primordial stage. Plant Breed 2002, 121, 370–377. [Google Scholar]
- Okazaki, K.; Kurimoto, K.; Miyajima, I.; Enami, A.; Mizuochi, H.; Matsumoto, Y.; Ohya, H. Induction of 2n pollen in tulips by arresting meiotic process with nitrous oxide gas. Euphytica 2005, 143, 101–114. [Google Scholar]
- Akutsu, M.; Kitamura, S.; Toda, R.; Miyajima, I.; Okazaki, K. Production of 2n pollen of Asiatic hybrid lilies by nitrous oxide treatment. Euphytica 2007, 155, 143–152. [Google Scholar]
- Nukui, S.; Kitamura, S.; Hioki, T.; Ootsuka, H.; Miyoshi, K.; Satou, T.; Takatori, U.; Oomiya, T.; Okazaki, K. N2O induces mitotic polyploidization in anther somatic cells and restores fertility in sterile interspecific hybrid lilies. Breed. Sci 2011, 61, 327–337. [Google Scholar]
- Kitamura, S.; Akutsu, M.; Okazaki, K. Mechanism of action of nitrous oxide gas applied as a polyploidizing agent during meiosis in lilies. Sex. Plant Reprod 2009, 22, 9–14. [Google Scholar]
- Barba-Gonzalez, R.; Miller, C.T.; Ramanna, M.S.; van Tuyl, J.M. Nitrous oxide (N2O) induces 2n gametes in sterile F1 hybrids between Oriental × Asiatic lily (Lilium) hybrids and leads to intergenomic recombination. Euphytica 2006, 148, 308–309. [Google Scholar]
- Karpechenko, G.D. The production of polyploidy gametes in hybrids. Hereditas 1927, 9, 349–368. [Google Scholar]
- Veilleux, R. Diploid and polyploid gametes in crop plants: Mechanisms of formation and utilization in plant breeding. Plant Breed. Rev 1983, 3, 253–288. [Google Scholar]
- Ramanna, M.S.; Jacobsen, E. Relevance of sexual polyploidization for crop improvement—A review. Euphytica 2003, 113, 3–18. [Google Scholar]
- Barba-Gonzalez, R.; Lim, K.B.; Zhou, S.; Ramanna, M.S.; van Tuyl, J.M. Interspecific hybridization in lily: The use of 2n gametes in interspecific lily hybrids. Floric. Ornam. Plant Biotech 2008, 5, 138–145. [Google Scholar]
- Lim, K.B.; Ramanna, M.S.; de Jong, J.H.; Jacobsen, E.; van Tuyl, J.M. Indeterminate meiotic restitution (IMR): A novel type of meiotic nuclear restitution mechanism detected in interspecific lily hybrids by GISH. Theor. Appl. Genet 2001, 103, 219–230. [Google Scholar]
- Asano, Y. Fertility of a hybrid between distantly related species in Lilium. Cytologia 1984, 49, 447–456. [Google Scholar]
- Van Tuyl, J.M.; Lim, K.B. Interspecific hybridization and polyploidization as tools in Ornamental plant breeding. Acta Hort 2003, 612, 13–22. [Google Scholar]
- Barba-Gonzalez, R.; Lokker, A.C.; Lim, K.B.; Ramanna, M.S.; van Tuyl, J.M. Use of 2n gametes for the production of sexual polyploids from sterile Oriental × Asiatic hybrids of lilies (Lilium). Theor. Appl. Genet 2004, 109, 1125–1132. [Google Scholar]
- Khan, N.; Zhou, S.; Ramanna, M.S.; Arens, P.; Herrera, J.; Visser, R.G.F.; van Tuyl, J.M. Potential for analytic breeding in allopolyploids: An illustration from Longiflorum × Asiatic hybrid lilies (Lilium). Euphytica 2009, 166, 399–409. [Google Scholar]
- Rieseberg, L.H.; Blackman, B.K. Speciation genes in plants. Ann. Bot 2010, 106, 439–455. [Google Scholar]
- Mayfield, D.; Chen, Z.J.; Pires, J.C. Epigenetic regulation of flowering time in polyploids. Curr. Opin. Plant Biol 2011, 14, 174–178. [Google Scholar]
- Nishiyama, I.; Yabuno, T. Causal relationships between the polar nuclei in double fertilization and interspecific cross-incompatibility in Avena. Cytologia 1978, 43, 453–466. [Google Scholar]
- Johnston, S.A.; Hanneman, R.E., Jr. Manipulations of endosperm balance number overcome crossing barriers between diploid solanum species. Science 1982, 217, 446–448. [Google Scholar]
- Kinoshita, T. Reproductive barrier and genomic imprinting in the endosperm of flowering plants. Genes Genet. Syst 2007, 82, 177–186. [Google Scholar]
- Martienssen, R.A. Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytol 2010, 186, 46–53. [Google Scholar]
- Köhler, C.; Mittelsten Scheid, O.; Erilova, A. The impact of the triploid block on the origin and evolution of polyploidy plants. Trends Genet 2010, 26, 142–148. [Google Scholar]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16, S154–S169. [Google Scholar]
- Leino, M.; Landgren, M.; Glimelius, K. Alloplasmic effects on mitochondrial transcriptional activity and RNA turnover result in accumulated transcripts of Arabidopsis orfs in cytoplasmc male-sterile Brassica napus. Plant J 2005, 42, 469–480. [Google Scholar]
- Köhler, C.; Wolff, P.; Spillane, C. Epigenetic mechanisms underlying genomic imprinting in plants. Annu. Rev. Plant Biol 2012, 63, 331–352. [Google Scholar]
- Kinoshita, T.; Yadegari, R.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 1999, 11, 1945–1952. [Google Scholar]
- Jullien, P.E.; Kinoshita, T.; Ohad, N.; Berger, F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 2006, 18, 1360–1372. [Google Scholar]
- Köhler, C.; Page, D.R.; Gagliardini, V.; Grossniklaus, U. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet 2005, 37, 28–30. [Google Scholar]
- Haig, D.; Westoby, M. Genomic imprinting in endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Phil. Trans. R. Soc. Lond. B 1991, 333, 1–13. [Google Scholar]
- Costa, L.M.; Yuan, J.; Rouster, J.; Paul, W.; Dickinson, H.; Gutierrez-Marcos, J.F. Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol 2012, 22, 160–165. [Google Scholar]
- Josefsson, C.; Dilkes, B.; Comai, L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol 2006, 16, 1322–1328. [Google Scholar]
- Erilova, A.; Brownfield, L.; Exner, V.; Rosa, M.; Twell, D.; Mittelsten Scheid, O.; Hennig, L.; Köhler, C. Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet 2009, 5, e1000663. [Google Scholar]
- Walia, H.; Josefsson, C.; Dilkes, B.; Kirkbride, R.; Harada, J.; Comai, L. Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol 2009, 19, 1128–1132. [Google Scholar]
- de Folter, S.; Immink, R.G.; Kieffer, M.; Parenicová, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar]
- Kang, I.H.; Steffen, J.G.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008, 20, 635–647. [Google Scholar]
- Ishikawa, R.; Ohnishi, T.; Kinoshita, Y.; Eiguchi, M.; Kurata, N.; Kinoshita, T. Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 2011, 65, 798–806. [Google Scholar]
- Kubo, T.; Newton, K.J. Angiosperm mitochondrial genomes and mutations. Mitochondrion 2008, 8, 5–14. [Google Scholar]
- Schnable, P.S.; Wise, R.P. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 1998, 3, 175–180. [Google Scholar]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci 2008, 13, 663–670. [Google Scholar]
- Bentolila, S.; Alfonso, A.A.; Hanson, M.R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 2002, 99, 10887–10892. [Google Scholar]
- Brown, G.G.; Formanova, N.; Jin, H.; Wargachuk, R.; Dendy, C.; Patil, P.; Laforest, M.; Zhang, J.; Cheung, W.Y.; Landry, B.S. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 2003, 35, 262–272. [Google Scholar]
- Koizuka, N.; Imai, R.; Fujimoto, H.; Hayakawa, T.; Kimura, Y.; Kohno-Murase, J.; Sakai, T.; Kawasaki, S.; Imamura, J. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 2003, 34, 407–415. [Google Scholar]
- Kazama, T.; Toriyama, K. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett 2003, 544, 99–102. [Google Scholar]
- Klein, R.R.; Klein, P.E.; Mullet, J.E.; Minx, P.; Rooney, W.L.; Schertz, K.F. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor. Appl. Genet 2005, 111, 994–1012. [Google Scholar]
- Wang, Z.; Zou, Y.; Li, X.; Zhang, Q.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D.; et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 2006, 18, 676–687. [Google Scholar]
- Gillman, J.D.; Bentolila, S.; Hanson, M.R. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J 2007, 49, 217–227. [Google Scholar]
- Kazama, T.; Nakamura, T.; Watanabe, M.; Sugita, M.; Toriyama, K. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J 2008, 55, 619–628. [Google Scholar]
- Uyttewaal, M.; Arnal, N.; Quadrado, M.; Martin-Canadell, A.; Vrielynck, N.; Hiard, S.; Gherbi, H.; Bendahmane, A.; Budar, F.; Mireau, H. Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. Plant Cell 2008, 20, 3331–3345. [Google Scholar]
- Cui, X.; Wise, R.P.; Schnable, P.S. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 1996, 272, 1334–1336. [Google Scholar]
- Fujii, S.; Toriyama, K. Suppressed expression of Retrograde-Regulated Male Sterility restores pollen fertility in cytoplasmic male sterile rice plants. Proc. Natl. Acad. Sci. USA 2009, 106, 9513–9518. [Google Scholar]
- Itabashi, E.; Iwata, N.; Fujii, S.; Kazama, T.; Toriyama, K. The fertility restorer gene, Rf2, for Lead Rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J 2011, 65, 359–367. [Google Scholar]
- Liu, F.; Cui, X.; Horner, H.; Weiner, H.; Schnable, P.S. Mitochondrial aldehyde dehydrogenase activity is requires for male fertility in maize. Plant Cell 2001, 13, 1063–1078. [Google Scholar]
- Fujii, S.; Kazama, T.; Yamada, M.; Toriyama, K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomics 2010, 11, 209. [Google Scholar]
- Fujii, S.; Toriyama, K. Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility. Plant Cell Physiol 2008, 49, 1484–1494. [Google Scholar]
- Takayama, S.; Isogai, A. Self-incompatibility in plants. Annu. Rev. Plant Biol 2005, 56, 467–489. [Google Scholar]
- Fujimoto, R.; Nishio, T. Self-incompatibility. Adv. Bot. Res 2007, 45, 139–154. [Google Scholar]
- Shiba, H.; Kakizaki, T.; Iwano, M.; Tarutani, Y.; Watanabe, M.; Isogai, A.; Takayama, S. Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet 2006, 38, 297–299. [Google Scholar]
- Tarutani, Y.; Shiba, H.; Ito, T.; Kakizaki, T.; Suzuki, G.; Watanabe, M.; Isogai, A.; Takayama, S. Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 2010, 466, 983–986. [Google Scholar]
- Tochigi, T.; Udagawa, H.; Li, F.; Kitashiba, H.; Nishio, T. The self-compatibility mechanism in Brassica napus L. is applicable to F1 hybrid breeding. Theor. Appl. Genet. 2011, 123, 475–482. [Google Scholar]
- Kimura, R.; Sato, K.; Fujimoto, R.; Nishio, T. Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleracea and Brassica rapa. Plant J 2002, 29, 215–223. [Google Scholar]
- Fujimoto, R.; Sugimura, T.; Fukai, E.; Nishio, T. Suppression of gene expression of a recessive SP11/SCR allele by an untranscribed SP11/SCR allele in Brassica self-incompatibility. Plant Mol. Biol 2006, 61, 577–587. [Google Scholar]
- Nasrallah, J.B.; Liu, P.; Sherman-Broyles, S.; Schmidt, R.; Nasrallah, M.E. Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics 2007, 175, 1965–1973. [Google Scholar]
- Goldberg, E.E.; Kohn, J.R.; Lande, R.; Robertson, K.A.; Smith, S.A.; Igic, B. Species selection maintains self-incompatibility. Science 2010, 330, 493–495. [Google Scholar]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar]
- Mestiri, I.; Chagué, V.; Tanguy, A.M.; Huneau, C.; Huteau, V.; Belcram, H.; Coriton, O.; Chalhoub, B.; Jahier, J. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol 2010, 186, 86–101. [Google Scholar]
- Song, K.; Lu, P.; Tang, K.; Osborn, T.C. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 7719–7723. [Google Scholar]
- Xiong, Z.; Gaeta, R.T.; Pires, J.C. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 2011, 108, 7908–7913. [Google Scholar]
- Szadkowski, E.; Eber, F.; Huteau, V.; Lodé, M.; Huneau, C.; Belcram, H.; Coriton, O.; Manzanares-Dauleux, M.J.; Delourme, R.; King, G.J.; et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol 2010, 186, 102–112. [Google Scholar]
- Szadkowski, E.; Eber, F.; Huteau, V.; Lodé, M.; Coriton, O.; Jenczewski, E.; Chévre, A.M. Polyploid formation pathways have an impact on genetic rearrangements in resynthesized Brassica napus. New Phytol 2011, 191, 884–894. [Google Scholar]
- Cifuentes, M.; Grandont, L.; Moore, G.; Chévre, A.M.; Jenczewski, E. Genetic regulation of meiosis in polyploid species: New insights into an old question. New Phytol 2010, 186, 29–36. [Google Scholar]
- Pontes, O.; Neves, N.; Silva, M.; Lewis, M.S.; Madlung, A.; Comai, L.; Viegas, W.; Pikaard, C.S. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 2004, 101, 18240–18245. [Google Scholar]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar]
- Wang, J.; Tian, L.; Madlung, A.; Lee, H.S.; Chen, M.; Lee, J.J.; Watson, B.; Kagochi, T.; Comai, L.; Chen, Z.J. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 2004, 167, 1961–1973. [Google Scholar]
- Xu, Y.; Zhong, L.; Wu, X.; Fang, X.; Wang, J. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 2009, 229, 471–483. [Google Scholar]
- Kakutani, T. Epi-alleles in plants: Inheritance of epigenetic information over generations. Plant Cell Physiol 2002, 43, 1106–1111. [Google Scholar]
- Fujimoto, R.; Sasaki, T.; Inoue, H.; Nishio, T. Hypomethylation and transcriptional reactivation of retrotransposon-like sequences in ddm1 transgenic plants of Brassica rapa. Plant Mol. Biol 2008, 66, 463–473. [Google Scholar]
- Tsukahara, S.; Kobayashi, A.; Kawabe, A.; Mathieu, O.; Miura, A.; Kakutani, T. Bursts of retrotransposition reproduced in Arabidopsis. Nature 2009, 461, 423–426. [Google Scholar]
- Sasaki, T.; Fujimoto, R.; Kishitani, S.; Nishio, T. Analysis of target sequences of DDM1s in Brassica rapa by MSAP. Plant Cell Rep 2011, 30, 81–88. [Google Scholar]
- Preuss, S.; Pikaard, C.S. rRNA gene silencing and nucleolar dominance: Insights into a chromosome-scale epigenetic on/off switch. Biochim. Biophys. Acta 2007, 1769, 383–392. [Google Scholar]
- Chen, Z.J.; Comai, L.; Pikaard, C.S. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. USA 1998, 95, 14891–14896. [Google Scholar]
- Lewis, M.S.; Pikaard, D.J.; Nasrallah, M.; Doelling, J.H.; Pikaard, C.S. Locus-specific ribosomal RNA gene silencing in nucleolar dominance. PLoS One 2007, 2, e815. [Google Scholar]
- Neves, N.; Heslop-Harrison, J.S.; Viegas, W. rRNA gene activity and control of expression mediated by methylation and imprinting during embryo development in wheat x rye hybrids. Theor. Appl. Genet 1995, 91, 529–533. [Google Scholar]
- Chen, Z.J.; Pikaard, C.S. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev 1997, 11, 2124–2136. [Google Scholar]
- Lawrence, R.J.; Earley, K.; Pontes, O.; Silva, M.; Chen, Z.J.; Neves, N.; Viegas, W.; Pikaard, C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 2004, 13, 599–609. [Google Scholar]
- Pontes, O.; Lawrence, R.J.; Silva, M.; Preuss, S.; Costa-Nunes, P.; Earley, K.; Neves, N.; Viegas, W.; Pikaard, C.S. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS One 2007, 2, e1157. [Google Scholar]
- Earley, K.; Lawrence, R.J.; Pontes, O.; Reuther, R.; Enciso, A.J.; Silva, M.; Neves, N.; Gross, M.; Viegas, W.; Pikaard, C.S. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 2006, 20, 1283–1293. [Google Scholar]
- Preuss, S.B.; Costa-Nunes, P.; Tucker, S.; Pontes, O.; Lawrence, R.J.; Mosher, R.; Kasschau, K.D.; Carrington, J.C.; Baulcombe, D.C.; Viegas, W.; et al. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell 2008, 32, 673–684. [Google Scholar]
- Ha, M.; Lu, J.; Tian, L.; Ramachandran, V.; Kasschau, K.D.; Chapman, E.J.; Carrington, J.C.; Chen, X.; Wang, X.J.; Chen, Z.J. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 2009, 106, 17835–17840. [Google Scholar]
- Kenan-Eichler, M.; Leshkowitz, D.; Tal, L.; Noor, E.; Melamed-Bessudo, C.; Feldman, M.; Levy, A.A. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 2011, 188, 263–272. [Google Scholar]
- Ng, D.W.; Zhang, C.; Miller, M.; Palmer, G.; Whiteley, M.; Tholl, D.; Chen, Z.J. cis- and trans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allotetraploids. Plant Cell 2011, 23, 1729–1740. [Google Scholar]
- Chague, V.; Just, J.; Mestri, I.; Balzergue, S.; Tanguy, A.M.; Huneau, C.; Huteau, V.; Blacramm, H.; Coriton, O.; Jahier, J.; Chalhoub, B. Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol 2010, 187, 1181–1194. [Google Scholar]
- Akhunova, A.R.; Matnivazov, R.T.; Liang, H.; Akhunov, E.D. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 2010, 11, 505. [Google Scholar]
- Qi, B.; Huang, W.; Zhu, B.; Zhong, X.; Guo, J.; Zhao, N.; Xu, C.; Zhang, H.; Pang, J.; Han, F.; Liu, B. Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol 2012, 10, 3. [Google Scholar]
- Hovav, R.; Udall, J.A.; Chaudhary, B.; Rapp, R.; Flagel, L.; Wendel, J.F. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. USA 2008, 105, 6191–6195. [Google Scholar]
- Udall, J.A.; Swanson, J.M.; Nettleton, D.; Percifield, R.J.; Wendel, J.F. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics 2006, 173, 1823–1827. [Google Scholar]
- Flagel, L.; Udall, J.; Nettleton, D.; Wendel, J. Duplicated gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 2008, 6, 16. [Google Scholar]
- Flagel, L.E.; Wendel, J.F. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol 2010, 186, 184–193. [Google Scholar]
- Zhuang, Y.; Chen, J.F. Changes of gene expression in early generations of the synthetic allotetraploid Cucumis x hytivus Chen et Kirkbride. Genet. Resour. Crop Evol 2009, 56, 1071–1076. [Google Scholar]
- Mudge, S.R.; Osabe, K.; Casu, R.E.; Bonnett, G.D.; Manners, J.M.; Birch, R.G. Efficient silencing of reporter transgenes coupled to known functional promoters in sugarcane, a highly polyploid crop species. Planta 2009, 229, 549–558. [Google Scholar]
- Adams, K.L.; Cronn, R.; Percifield, R.; Wendel, J.F. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 2003, 100, 4649–4654. [Google Scholar]
- Dong, S.W.; Adams, K.L. Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol 2011, 190, 1045–1057. [Google Scholar]
- Anssour, S.; Baldwin, I.T. Variation in antiherbivore defense responses in synthetic Nicotiana allopolyploids correlates with changes in uniparental patterns of gene expression. Plant Physiol 2010, 153, 1907–1918. [Google Scholar]
- Buggs, R.J.; Elliott, N.M.; Zhang, L.; Koh, J.; Viccini, L.F.; Soltis, D.E.; Soltis, P.S. Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol 2010, 186, 175–183. [Google Scholar]
- Marmagne, A.; Brabant, P.; Thiellement, H.; Alix, K. Analysis of gene expression in resynthesized Brassica napus allotetraploids: Transcriptional changes do not explain differential protein regulation. New Phytol 2010, 186, 216–227. [Google Scholar]
- Kong, F.; Mao, S.; Jiang, J.; Wang, J.; Fang, X.; Wang, Y. Proteomic changes in newly synthesized Brassica napus allotetraploids and their early generations. Plant Mol. Biol. Rep 2011, 29, 927–935. [Google Scholar]
- Pont, C.; Murat, F.; Confolent, C.; Balzergue, S.; Salse, J. RNA-seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.). Genome Biol 2011, 12, R119. [Google Scholar]
- Zhang, Y.; Xu, G.H.; Guo, X.Y.; Fan, L.J. Two ancient rounds of polyploidy in rice genome. J. Zhejiang Univ. Sci. B 2005, 6, 87–90. [Google Scholar]
- Throude, M.; Bolot, S.; Bosio, M.; Pont, C.; Sarda, X.; Quraishi, U.M.; Bourgis, F.; Lessard, P.; Rogowsky, P.; Ghesquiere, A.; et al. Structure and expression analysis of rice paleo duplications. Nucleic Acids Res 2009, 37, 1248–1259. [Google Scholar]
- Lackey, E.; Ng, D.W.K.; Chen, Z.J. RNAi-mediated down-regulation of DCL1 and AGO1 induces developmental changes in resynthesized Arabidopsis allotetraploids. New Phytol 2010, 186, 207–215. [Google Scholar]
- Osabe, K.; Mudge, S.R.; Graham, M.W.; Birch, R.G. RNAi mediated down-regulation of PDS gene expression in Sugarcane (Saccharum), a highly polyploid crop. Trop. Plant Biol 2009, 2, 143–148. [Google Scholar]




© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Osabe, K.; Kawanabe, T.; Sasaki, T.; Ishikawa, R.; Okazaki, K.; Dennis, E.S.; Kazama, T.; Fujimoto, R. Multiple Mechanisms and Challenges for the Application of Allopolyploidy in Plants. Int. J. Mol. Sci. 2012, 13, 8696-8721. https://doi.org/10.3390/ijms13078696
Osabe K, Kawanabe T, Sasaki T, Ishikawa R, Okazaki K, Dennis ES, Kazama T, Fujimoto R. Multiple Mechanisms and Challenges for the Application of Allopolyploidy in Plants. International Journal of Molecular Sciences. 2012; 13(7):8696-8721. https://doi.org/10.3390/ijms13078696
Chicago/Turabian StyleOsabe, Kenji, Takahiro Kawanabe, Taku Sasaki, Ryo Ishikawa, Keiichi Okazaki, Elizabeth S. Dennis, Tomohiko Kazama, and Ryo Fujimoto. 2012. "Multiple Mechanisms and Challenges for the Application of Allopolyploidy in Plants" International Journal of Molecular Sciences 13, no. 7: 8696-8721. https://doi.org/10.3390/ijms13078696



