Quantitative Expression of C-Type Lectin Receptors in Humans and Mice
Abstract
:1. Introduction
2. Results and Discussion
2.1. C-Type Lectin Receptor mRNA Expression in Adult Human Tissues
2.2. C-Type Lectin Receptor mRNA Expression in Adult Murine Tissues
2.3. C-Type Lectin Receptor mRNA Expression in Ischemia-Reperfusion Injury (IRI)
2.4. C-Type Lectin Receptor mRNA Expression in Progressive Renal Fibrosis
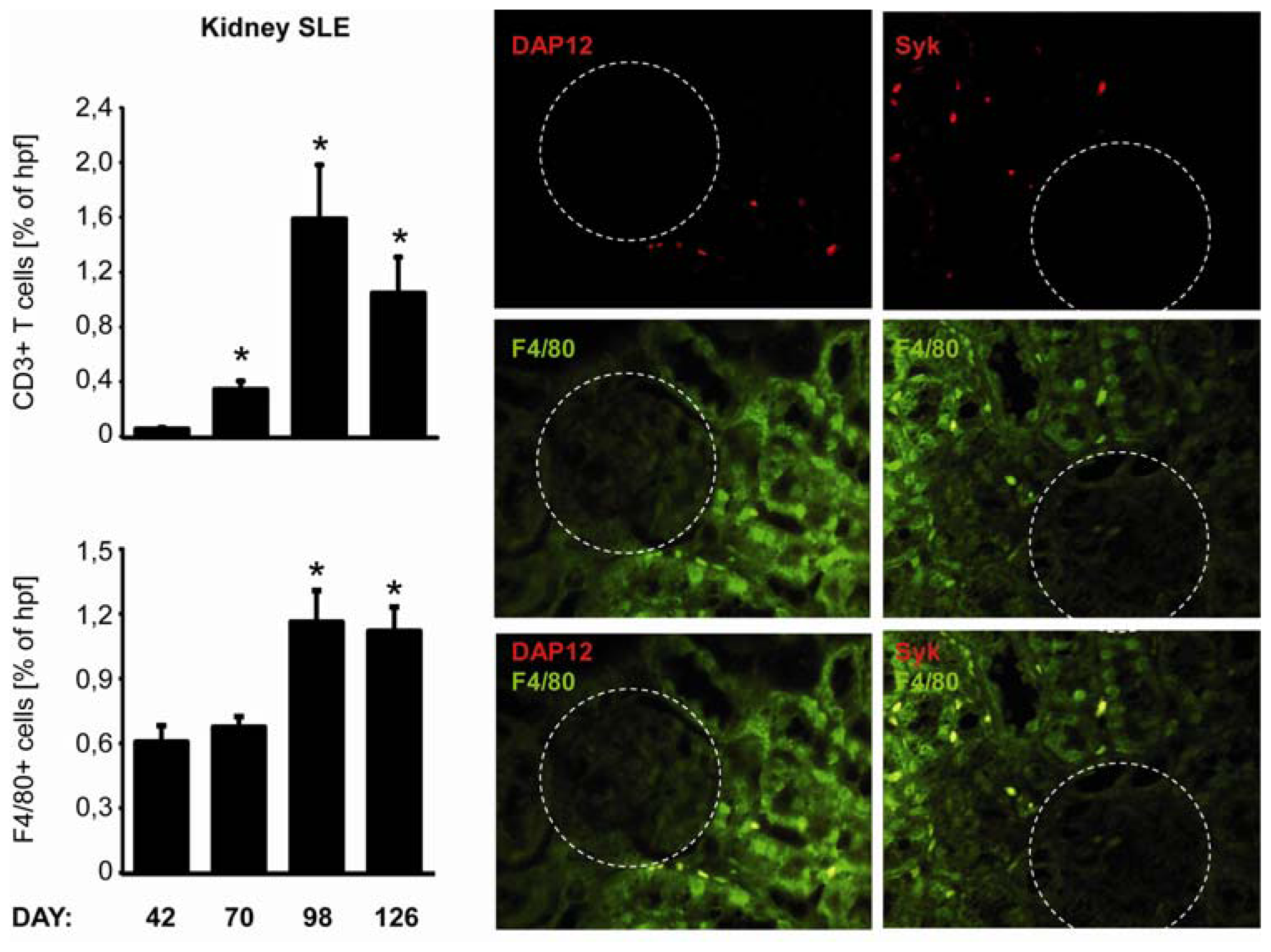
2.5. C-Type Lectin Receptor mRNA Expression in Systemic Autoimmunity and Immune Complex Glomerulonephritis
2.6. Role of C-Type Lectin Receptors in Tissue Homeostasis
3. Experimental Section
3.1. Animal Studies
3.2. Human Solid Organ cDNA
3.3. Quantitative Real-Time RT-PCR
3.4. Morphological Evaluation
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
ijms-13-10113-s001.pdfAcknowledgments
References
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar]
- Osorio, F.; Reis e Sousa, C. Myeloid C-type lectin receptors in pathogen recognition and host defense. Immunity 2011, 34, 651–664. [Google Scholar]
- Geijtenbeek, T.B.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol 2009, 9, 465–479. [Google Scholar]
- Schreibelt, G.; Klinkenberg, L.J.; Cruz, L.J.; Tacken, P.J.; Tel, J.; Kreutz, M.; Adema, G.J.; Brown, G.D.; Figdor, C.G.; de Vries, I.J. The C-type lectin receptor CLEC9A mediates antigen uptake and (cross-) presentation by human blood BDCA3+ myeloid dendritic cells. Blood 2012, 119, 2284–2292. [Google Scholar]
- Kimbrell, D.A.; Beutler, B. The evolution and genetics of innate immunity. Nat. Rev. Genet 2001, 2, 256–267. [Google Scholar]
- Zhang, D.; Zhang, G.; Hayden, M.S.; Greenblatt, M.B.; Bussey, C.; Flavell, R.A.; Ghosh, S. A toll-like receptor that prevents infection by uropathogenic bacteria. Science 2004, 303, 1522–1526. [Google Scholar]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar]
- Wagner, H. The immunobiology of the TLR9 subfamily. Trends Immunol 2004, 25, 381–386. [Google Scholar]
- Caminschi, I.; Corbett, A.J.; Zahra, C.; Lahoud, M.; Lucas, K.M.; Sofi, M.; Vremec, D.; Gramberg, T.; Pohlmann, S.; Curtis, J.; et al. Functional comparison of mouse CIRE/mouse DC-SIGN and human DC-SIGN. Int. Immunol 2006, 18, 741–753. [Google Scholar]
- Hornung, V.; Rothenfusser, S.; Britsch, S.; Krug, A.; Jahrsdorfer, B.; Giese, T.; Endres, S.; Hartmann, G. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol 2002, 168, 4531–4537. [Google Scholar]
- Zarember, K.A.; Godowski, P.J. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J. Immunol 2002, 168, 554–561. [Google Scholar]
- Lassen, S.; Lech, M.; Rommele, C.; Mittruecker, H.W.; Mak, T.W.; Anders, H.J. Ischemia reperfusion induces IFN regulatory factor 4 in renal dendritic cells, which suppresses postischemic inflammation and prevents acute renal failure. J. Immunol 2010, 185, 1976–1983. [Google Scholar]
- Anders, H.J; Ryu, M. Renal microenvironments and macrophage phenotypes determine progression or resolution of renal inflammation and fibrosis. Kidney Int. 2011, 80, 915–925. [Google Scholar]
- Patole, P.S.; Pawar, R.D.; Lech, M.; Zecher, D.; Schmidt, H.; Segerer, S.; Ellwart, A.; Henger, A.; Kretzler, M.; Anders, H.J. Expression and regulation of Toll-like receptors in lupus-like immune complex glomerulonephritis of MRL-Fas(lpr) mice. Nephrol. Dial. Transplant 2006, 21, 3062–3073. [Google Scholar]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar]
- Kono, H.; Rock, K.L. How dying cells alert the immune system to danger. Nat. Rev. Immunol 2008, 8, 279–289. [Google Scholar]
- Van Vliet, S.J.; den Dunnen, J.; Gringhuis, S.I.; Geijtenbeek, T.B.; van Kooyk, Y. Innate signaling and regulation of Dendritic cell immunity. Curr. Opin. Immunol 2007, 19, 435–440. [Google Scholar]
- Lech, M.; Avila-Ferrufino, A.; Skuginna, V.; Susanti, H.E.; Anders, H.J. Quantitative expression of RIG-like helicase, NOD-like receptor and inflammasome-related mRNAs in humans and mice. Int. Immunol 2010, 22, 717–728. [Google Scholar]
- Kronin, V.; Wu, L.; Gong, S.; Nussenzweig, M.C.; Shortman, K. DEC-205 as a marker of dendritic cells with regulatory effects on CD8 T cell responses. Int. Immunol 2000, 12, 731–735. [Google Scholar]
- Ferwerda, B.; Ferwerda, G.; Plantinga, T.S.; Willment, J.A.; van Spriel, A.B.; Venselaar, H.; Elbers, C.C.; Johnson, M.D.; Cambi, A.; Huysamen, C.; et al. Human dectin-1 deficiency and mucocutaneous fungal infections. N. Engl. J. Med 2009, 361, 1760–1767. [Google Scholar]
- Kock, G.; Bringmann, A.; Held, S.A.; Daecke, S.; Heine, A.; Brossart, P. Regulation of dectin-1-mediated dendritic cell activation by peroxisome proliferator-activated receptor-gamma ligand troglitazone. Blood 2011, 117, 3569–3574. [Google Scholar]
- Gross, O.; Gewies, A.; Finger, K.; Schafer, M.; Sparwasser, T.; Peschel, C.; Forster, I.; Ruland, J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 2006, 442, 651–656. [Google Scholar]
- Gringhuis, S.I.; den Dunnen, J.; Litjens, M.; van der Vlist, M.; Wevers, B.; Bruijns, S.C.; Geijtenbeek, T.B. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-κB activation through Raf-1 and Syk. Nat. Immunol 2009, 10, 203–213. [Google Scholar]
- Mocsai, A.; Ruland, J.; Tybulewicz, V.L. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat. Rev. Immunol 2010, 10, 387–402. [Google Scholar]
- Bjorge, J.D.; Jakymiw, A.; Fujita, D.J. Selected glimpses into the activation and function of Src kinase. Oncogene 2000, 19, 5620–5635. [Google Scholar]
- Lopez-Guisa, J.M.; Cai, X.; Collins, S.J.; Yamaguchi, I.; Okamura, D.M.; Bugge, T.H.; Isacke, C.M.; Emson, C.L.; Turner, S.M.; Shankland, S.J.; et al. Mannose receptor 2 attenuates renal fibrosis. J. Am. Soc. Nephrol 2012, 23, 236–251. [Google Scholar]
- Elola, M.T.; Chiesa, M.E.; Alberti, A.F.; Mordoh, J.; Fink, N.E. Galectin-1 receptors in different cell types. J. Biomed. Sci 2005, 12, 13–29. [Google Scholar]
- Camby, I.; Le Mercier, M.; Lefranc, F.; Kiss, R. Galectin-1: A small protein with major functions. Glycobiology 2006, 16, 137R–157R. [Google Scholar]
- Gingras, M.C.; Lapillonne, H.; Margolin, J.F. TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol. Immunol 2002, 38, 817–824. [Google Scholar]
- Weidenbusch, M.; Anders, H.J. Tissue Microenvironments define and get reinforced by macrophage phenotypes in homeostasis or during inflammation, repair and fibrosis. J. Innate Immun 2012. [Google Scholar] [CrossRef]
- Lech, M.; Garlanda, C.; Mantovani, A.; Kirschning, C.J.; Schlondorff, D.; Anders, H.J. Different roles of TiR8/Sigirr on toll-like receptor signaling in intrarenal antigen-presenting cells and tubular epithelial cells. Kidney Int 2007, 72, 182–192. [Google Scholar]
- Lech, M.; Avila-Ferrufino, A.; Allam, R.; Segerer, S.; Khandoga, A.; Krombach, F.; Garlanda, C.; Mantovani, A.; Anders, H.J. Resident dendritic cells prevent postischemic acute renal failure by help of single Ig IL-1 receptor-related protein. J. Immunol 2009, 183, 4109–4118. [Google Scholar]
- Skuginna, V.; Lech, M.; Allam, R.; Ryu, M.; Clauss, S.; Susanti, H.E.; Rommele, C.; Garlanda, C.; Mantovani, A.; Anders, H.J. Toll-like receptor signaling and SIGIRR in renal fibrosis upon unilateral ureteral obstruction. PLoS One 2011, 6, e19204. [Google Scholar]
- Kulkarni, O.; Pawar, R.D.; Purschke, W.; Eulberg, D.; Selve, N.; Buchner, K.; Ninichuk, V.; Segerer, S.; Vielhauer, V.; Klussmann, S.; et al. Spiegelmer inhibition of CCL2/MCP-1 ameliorates lupus nephritis in MRL-(Fas)lpr mice. J. Am. Soc. Nephrol 2007, 18, 2350–2358. [Google Scholar]







| GAPDH | Actin | α-tubulin | Phospholipase A2 | β-mikroglobulin | |
|---|---|---|---|---|---|
| N | 11 | 11 | 11 | 11 | 11 |
| GM | 20.35 | 17.97 | 18.98 | 20.55 | 16.49 |
| AM | 20.36 | 18.01 | 19.01 | 20.60 | 16.58 |
| Minimum | 19.12 | 17.35 | 17.96 | 18.39 | 13.71 |
| Maximum | 21.59 | 20.34 | 20.40 | 23.88 | 21.13 |
| SD | 0.75 | 1.17 | 1.12 | 1.49 | 1.84 |
| Variance | 0.56 | 1.36 | 1.26 | 2.21 | 3.38 |
| CV | 0.11 | 0.24 | 0.24 | 0.45 | 0.56 |
| GAPDH | Actin | α-tubulin | Phospholipase A2 | β-mikroglobulin | 18 s | |
|---|---|---|---|---|---|---|
| N | 11 | 11 | 11 | 11 | 11 | 11 |
| GM | 20.33 | 18.46 | 20.27 | 20.83 | 18.24 | 8.32 |
| AM | 20.34 | 18.49 | 20.29 | 20.88 | 18.37 | 8.40 |
| Minimum | 19.31 | 16.85 | 18.98 | 18.75 | 15.28 | 7.33 |
| Maximum | 21.38 | 20.01 | 21.28 | 23.84 | 21.07 | 11.22 |
| SD | 0.61 | 1.03 | 0.75 | 1.58 | 2.25 | 1.24 |
| Variance | 0.38 | 1.07 | 0.56 | 2.50 | 5.08 | 1.53 |
| CV | 0.08 | 0.20 | 0.11 | 0.52 | 0.93 | 0.13 |
| Spleen MRL | Kidney MRL | UUO | IRI | |
|---|---|---|---|---|
| N | 41 | 55 | 25 | 25 |
| GM | 9.38 | 7.86 | 9.22 | 9.02 |
| AM | 9.40 | 7.91 | 9.23 | 9.03 |
| Minimum | 8.10 | 6.44 | 8.36 | 7.95 |
| Maximum | 10.90 | 9.82 | 10.41 | 9.89 |
| SD | 0.70 | 0.88 | 0.52 | 0.56 |
| Variance | 0.48 | 0.77 | 0.27 | 0.31 |
| CV | 0.05 | 0.06 | 0.03 | 0.03 |
| Mouse right primer sequence | Mouse left primer sequence | Accession Nr. | Efficiency | Error value | |
|---|---|---|---|---|---|
| Dectin-1 | GTGCAGTAAGCTTTCCTGGG | TCCCGCAATCAGAGTGAAG | NM_020008 | 1.99 | 0.0007 |
| MR1 | GTGGATTGTCTTGTGGAGCA | TTGTGGTGAGCTGAAAGGTG | NM_008625 | 2.16 | 0.0140 |
| MR2 | CTCCAGACAGCCCTGCAT | GTCTTGCTTCTCGGGGGACT | NM_008626 | 1.81 | 0.0156 |
| DC-SIGN | CTGCACAGTCTTCCTCTCCC | TGGTACTGGGTAGATGGTTCA | NM_133238 | 2.00 | 0.0188 |
| Syk | TCTGCACCCCTTCAGAGTTC | TCCTTTCAACGTTCCATGCT | NM_011518 | 1.79 | 0.0510 |
| Card-9 | ATGAGGCTGTGCCTGAGC | GCTGCAAGGACGAGAACTATG | NM_001037747 | 2.08 | 0.0174 |
| Bcl-10 | TGCACGTAGATGATCAAAATGTC | ACGGAGGAGGATTTGACTGA | NM_009740 | 2.13 | 0.0740 |
| Malt-1 | CAAAAGGATGTCCAGTTGCC | CACACTGAGGTTCTTCCGCT | NM_172833 | 1.82 | 0.0065 |
| Src | TACCACTCCTCAGCCTGGAT | ACACGAGGAAGGTGGATGTC | NM_009271 | 2.19 | 0.0374 |
| Dec-205 | TTCAGACCAATCCACAACCA | AGCTCACCTACCCAGCTTCA | NM_013825 | 2.03 | 0.1900 |
| Galectin-1 | GCGAGGATTGAAGTGTAGGC | AATGTCTCAAAGTTCGGGGA | NM_008495 | 1.96 | 0.0085 |
| Tim-3 | AGCCCATGTGGAAATTTTTG | CTCCAAGAACCCTAACCACG | NM_134250 | 2.00 | 0.0145 |
| Trem-1 | CACTGTCAAAGTCTGGCCCT | ACTGCTGTGCGTGTTCTTTG | NM_021406 | 2.00 | 0.0740 |
| DAP-12 | TTGCCTCTGTGTGTTGAGGT | CGGAAACAACACATTGCTGA | NM_011662 | 1.90 | 0.0379 |
| Human right primer sequence | Human left primer sequence | Accession Nr | Efficiency | Error value | |
| Dectin-1 | GGAGATGGGTTTTCTTGGGT | GACTGAGGTACCATGGCTCTG | NM_022570 | 2.06 | 0.0541 |
| MR1 | CCCATCGGAATTTCTGTGAT | GGGTGCTGTTCTCCTACTGG | NM_002438 | 2.06 | 0.0587 |
| MR2 | CAGTCCATGGCTGAAGATGA | GCTGCGTCCTGCTCCTC | NM_006039 | 2.22 | 0.1030 |
| DC-SIGN | TTGTTGGGCTCTCCTCTGTT | AAGTAACCGCTTCACCTGGA | NM_021155 | 2.00 | 0.0122 |
| Syk | AAAGAAGGGCAGGTGGTTG | GAGAGCGAGGAGGAGCG | NM_003177 | 2.07 | 0.0197 |
| Card-9 | CTGTGCGTGCAGCTCCT | TCCAAGATGTACAAGGACCG | NM_052813 | 2.10 | 0.2160 |
| Bcl-10 | TTGCACGTAGATGATCAAAATGT | TCCCTCACCGAGGAGGAC | NM_003921 | 2.00 | 0.0178 |
| Malt-1 | GCCAAGACTGCCTTTGACTC | TTTCCTGCAGGCTATGGAAC | NM_006785 | 2.17 | 0.0364 |
| Src | TAGTTGCTGGGGATGTAGCC | CTGTCCTTCAAGAAAGGCGA | NM_005417 | 2.12 | 0.0134 |
| Dec-205 | CCAGCCAAAAACTTCTCATTT | TGGCTTCATGGGTCATGTTA | NM_002349 | 2.01 | 0.2010 |
| Galectin-1 | AGGTTGTTGCTGTCTTTGCC | CAAACCTGGAGAGTGCCTTC | NM_002305 | 1.87 | 0.0810 |
| Tim-3 | GCGAATTCCCTCTGCTACTG | CTTCGGCGCTTTAATTTTCA | NM_032782 | 2.02 | 0.0914 |
| Trem-1 | TACTCAGGAATCCACCAGCC | CCGATGTCTCCACTCCTGAC | NM_018643 | 1.92 | 0.0731 |
| DAP-12 | GTCATGATTCGGGCTCATTT | GAGACCGAGTCGCCTTATCA | NM_003332 | 2.08 | 0.0191 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lech, M.; Susanti, H.E.; Römmele, C.; Gröbmayr, R.; Günthner, R.; Anders, H.-J. Quantitative Expression of C-Type Lectin Receptors in Humans and Mice. Int. J. Mol. Sci. 2012, 13, 10113-10131. https://doi.org/10.3390/ijms130810113
Lech M, Susanti HE, Römmele C, Gröbmayr R, Günthner R, Anders H-J. Quantitative Expression of C-Type Lectin Receptors in Humans and Mice. International Journal of Molecular Sciences. 2012; 13(8):10113-10131. https://doi.org/10.3390/ijms130810113
Chicago/Turabian StyleLech, Maciej, Heni Eka Susanti, Christoph Römmele, Regina Gröbmayr, Roman Günthner, and Hans-Joachim Anders. 2012. "Quantitative Expression of C-Type Lectin Receptors in Humans and Mice" International Journal of Molecular Sciences 13, no. 8: 10113-10131. https://doi.org/10.3390/ijms130810113




