MUC16/CA125 in the Context of Modular Proteins with an Annotated Role in Adhesion-Related Processes: In Silico Analysis
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. Dataset
3.2. Similarity Search
3.3. Protein Function Prediction
3.4. CA125-Immunoreactivity
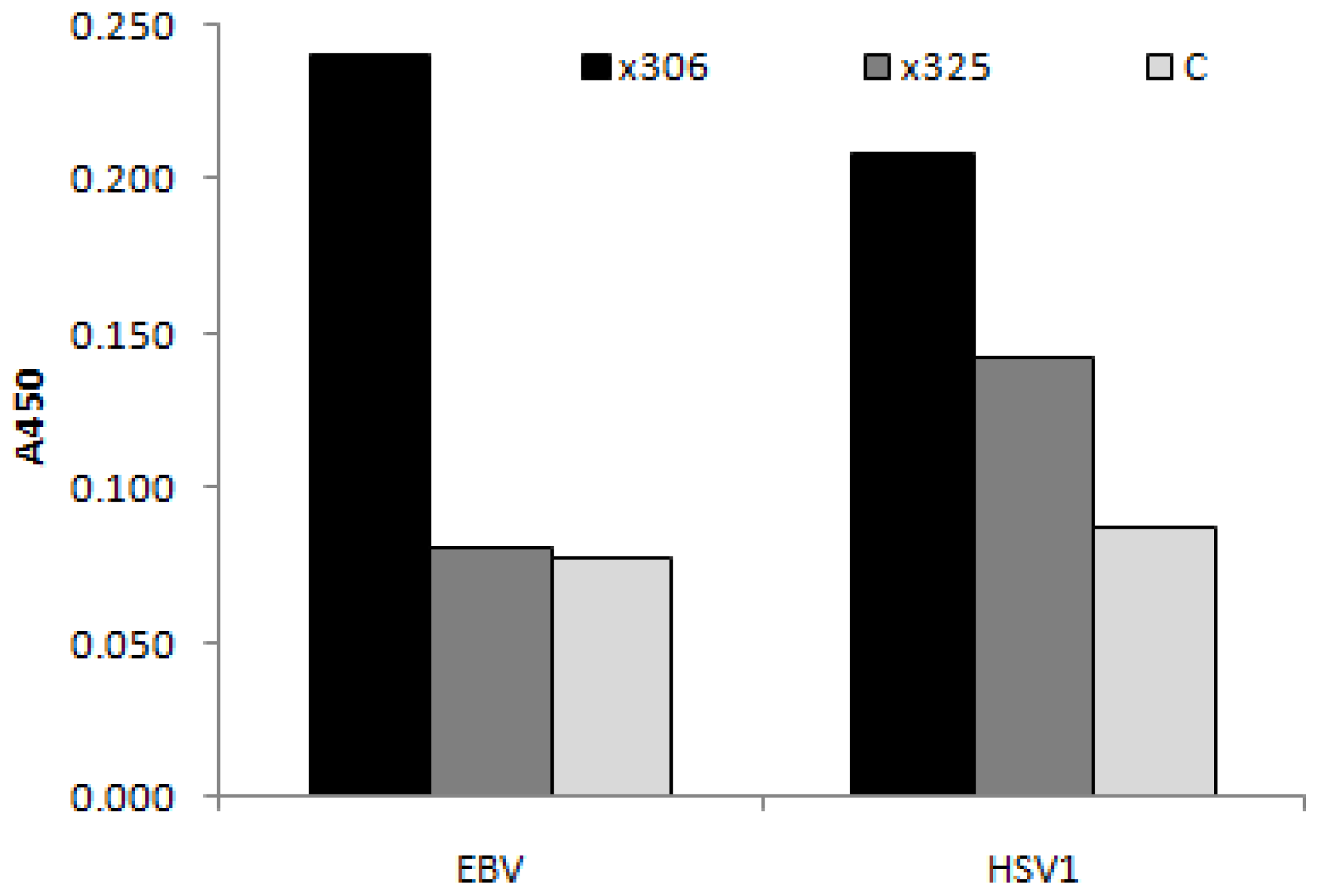
3.4.1. Viral Antigens
3.4.2. Crude Yeast (Saccharomyces cerevisiae) Extract
4. Conclusions
Acknowledgment
- Conflict of InterestThe authors declare no financial or commercial conflict of interest.
References
- Gum, J.R. Mucin genes and proteins they encode: Structure, diversity and regulation. Am. J. Respir. Cell Mol. Biol 1992, 7, 557–564. [Google Scholar]
- Perez-Vilar, J.; Hill, R.L. Mucin Family of Glycoproteins. In Encyclopedia of Biological Chemistry; Lennarz, L., Ed.; Academic Press/Elsevier: Oxford, UK, 2004; Volume 2, pp. 758–764. [Google Scholar]
- Desseyn, J.L.; Tetaert, D.; Gouyer, V. Architecture of the large membrane-bound mucins. Gene 2008, 410, 215–222. [Google Scholar]
- Hattrup, C.L.; Gendler, S.J. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol 2008, 70, 431–457. [Google Scholar]
- Jonckheere, N.; van Seuningen, I. The membrane-bound mucins: From cell signalling to transcriptional regulation and expression in epithelial cancers. Biochimie 2009, 92, 1–11. [Google Scholar]
- Bafna, S.; Kaur, S.; Batra, S.K. Membrane-Bound mucins: The mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 2010, 29, 2893–2904. [Google Scholar]
- Palileo, C.; Kaunitz, J.D. Gastrointestinal defense mechanisms. Curr. Opin. Gastroenterol 2011, 27, 543–548. [Google Scholar]
- McGuckin, M.A.; Lindén, S.K.; Sutton, P.; Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol 2011, 4, 265–278. [Google Scholar]
- Parker, D.; Prince, A. Innate immunity in the respiratory epithelium. Am. J. Respir. Cell Mol. Biol 2011, 45, 189–201. [Google Scholar]
- Montz, F.J. CA 125. In Serological Cancer Markers; Sell, S., Ed.; The Humana Press: Totowa, NJ, USA, 1992; pp. 417–425. [Google Scholar]
- Scholler, N.; Urban, N. CA 125 in ovarian cancer. Biomark. Med 2007, 1, 513–523. [Google Scholar]
- Perez, B.H.; Gipson, I.K. Focus on molecules: Human mucin MUC16. Exp. Eye Res 2008, 87, 400–401. [Google Scholar]
- Yin, B.W.; Lloyd, K.O. Molecular cloning of the CA 125 ovarian cancer antigen: Identification as a new mucin, MUC16. J. Biol. Chem 2001, 276, 27371–27375. [Google Scholar]
- O’Brien, T.J.; Beard, J.B.; Underwood, L.J.; Dennis, R.A.; Santin, A.D.; York, L. The CA 125 gene: An extracellular superstructure dominated by repeat sequences. Tumour Biol 2001, 22, 348–366. [Google Scholar]
- O’Brien, T.J.; Beard, J.B.; Underwood, L.J.; Shigemasa, K. The CA 125 gene: A newly discovered extension of the glycosylated N-terminal domain doubles the size of this extracellular superstructure. Tumour Biol 2002, 23, 154–169. [Google Scholar]
- Maeda, T.; Inoue, M.; Koshiba, S.; Yabuki, T.; Aoki, M.; Nunokawa, E.; Seki, E.; Matsuda, T.; Motoda, Y.; Kobayashi, A.; et al. Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA 125 (MUC16). J. Biol. Chem 2004, 279, 13174–13182. [Google Scholar]
- IPR000082 SEA. Available online: http://www.ebi.ac.uk/interpro/IEntry?ac=IPR000082 accessed on 3 April 2012.
- Sedgwick, S.G.; Smerdon, S.J. The ankyrin repeat: A diversity of interactions on a common structural framework. Trends Biochem. Sci 1999, 24, 311–316. [Google Scholar]
- Wong, N.K.; Easton, R.L.; Pancio, M.; Sutton-Smith, M.; Morrison, J.C.; Lattanzio, F.A.; Moris, H.R.; Clark, G.F.; Dell, A.; Patankar, M.S. Characterization of the oligosaccharides associated with human ovarian tumor marker CA125. J. Biol. Chem 2003, 278, 28619–28634. [Google Scholar]
- Jankovic, M.; Tapuskovic, B. Molecular forms and microheterogeneity of the oligosaccharide chains of pregnancy-associated CA125 antigen. Hum. Reprod 2005, 20, 2632–2638. [Google Scholar]
- Jankovic, M.; Milutinovic, B. Glycoforms of CA125 antigen as possible cancer biomarker. Cancer Biomark 2008, 1, 1–8. [Google Scholar]
- Jankovic, M.M.; Milutinovic, B.S. Pregnancy-Associated CA125 antigen as mucin: Evaluation of ferning morphology. Mol. Hum. Reprod 2007, 13, 405–408. [Google Scholar]
- Bouanène, H.; Saibi, W.; Mokni, M.; Sriha, B.; Ben Fatma, L.; Ben Limem, H.; Ben Ahmed, S.; Gargouri, A.; Miled, A. Biochemical and morphological differences between CA125 isolated from healthy women and patients with epithelial ovarian cancer from Tunisian population. Pathol. Oncol. Res 2012, 18, 325–330. [Google Scholar]
- Whittaker, G.R.; Wheldon, L.A.; Giles, L.E.; Stocks, J.M.; Halliburton, I.W.; Killington, R.A.; Meredith, D.M. Characterization of the high Mr glycoprotein (gp300) of equine herpesvirus type 1 as a novel glycoprotein with extensive O-linked carbohydrate. J. Gen. Virol 1990, 71, 2416. [Google Scholar]
- Wellington, J.E.; Allen, G.P.; Gooley, A.A.; Love, D.N.; Packer, N.H.; Yan, J.X.; Whalley, J.M. The highly O-glycosylated glycoprotein gp2 of equine herpesvirus 1 is encoded by gene 71. J. Virol. 1996, 70, 8198. [Google Scholar]
- Learmonth, G.S.; Love, D.N.; Gilkerson, J.R.; Wellington, J.E.; Whalley, J.M. The C-terminal regions of the envelope glycoprotein gp2 of equine herpesvirus 1 and 4 are antigenically distinct. Arch. Virol 2002, 147, 607–615. [Google Scholar]
- Tanner, J.; Weis, J.; Fearon, D.; Whang, Y.; Kieff, E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 1987, 50, 203–213. [Google Scholar]
- Janz, A.; Oezel, M.; Kurzeder, C.; Mautner, J.; Pich, D.; Kost, M.; Hammerschmidt, W.; Delecluse, H.J. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol 2000, 74, 10142–10152. [Google Scholar]
- Luo, B.; Liu, M.; Chao, Y.; Wang, Y.; Jing, Y.; Sun, Z. Characterization of Epstein-Barr virus gp350/220 gene variants in virus isolates from gastric carcinoma and nasopharyngeal carcinoma. Arch. Virol 2012, 157, 207–216. [Google Scholar]
- Cell wall surface anchor family protein. Available online: http://www.uniprot.org/uniprot/B2ISC7 accessed on 5 April 2012.
- Ding, F.; Tang, P.; Hsu, M.H.; Cui, P.; Hu, S.; Yu, J.; Chiu, C.H. Genome evolution driven by host adaptations results in a more virulent and antimicrobial-resistant Streptococcus pneumoniae serotype 14. BMC Genomics 2009, 10, 158. [Google Scholar]
- Serine-rich adhesin for platelets. Available online: http://www.uniprot.org/uniprot/Q4L9P0 accessed on 5 April 2012.
- Takeuchi, F.; Watanabe, S.; Baba, T.; Yuzawa, H.; Ito, T.; Morimoto, Y.; Kuroda, M.; Cui, L.; Takahashi, M.; Ankai, A.; et al. Whole-Genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol 2005, 187, 7292–7308. [Google Scholar]
- Cell surface flocculin. Available online: http://www.uniprot.org/uniprot/E9P8M0 accessed on 5 April 2012.
- Lo, W.S.; Dranginis, A.M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 1998, 9, 161–171. [Google Scholar]
- Karunanithi, S.; Vadaie, N.; Chavel, C.A.; Birkaya, B.; Joshi, J.; Grell, L.; Cullen, P.J. Shedding of the mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr. Biol 2010, 20, 1389–1395. [Google Scholar]
- Veelders, M.; Brückner, S.; Ott, D.; Unverzagt, C.; Mösch, H.U.; Essen, L.O. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 22511–22516. [Google Scholar]
- Muc1p. Available online: http://www.uniprot.org/uniprot/C8ZAR8 accessed on 6 April 2012.
- Lambrechts, M.G.; Bauer, F.F.; Marmur, J.; Pretorius, I.J. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 8419–8424. [Google Scholar]
- Secundino, N.; Kimblin, N.; Peters, N.C.; Lawyer, P.; Capul, A.A.; Beverley, S.M.; Turco, S.J.; Sacks, D. Proteophosphoglycan confers resistance of Leishmania major to midgut digestive enzymes induced by blood feeding in vector sand flies. Cell Microbiol 2010, 12, 906–918. [Google Scholar]
- Aebischer, T.; Harbecke, D.; Ilg, T. Proteophosphoglycan, a major secreted product of intracellular Leishmania mexicana amastigotes, is a poor b-cell antigen and does not elicit a specific conventional CD4+ T-cell response. Infect. Immun 1999, 67, 5379–5385. [Google Scholar]
- Marchler-Bauer, A.; Shennan, L.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acid Res 2011, 39, 225–229. [Google Scholar]
- Hsieh, W.C.; Chang, Y.; Hsu, M.C.; Lan, B.S.; Hsiao, G.C.; Chuang, H.C.; Su, I.J. Emergence of anti-red blood cell antibodies triggers red cell phagocytosis by activated macrophages in a rabbit model of Epstein-Barr virus-associated hemophagocytic syndrome. Am. J. Pathol 2007, 170, 1629–1639. [Google Scholar]
- Bairey, O.; Blickstein, D.; Stark, P.; Prokocimer, M.; Nativ, H.M.; Kirgner, I.; Shaklai, M. Serum CA 125 as a prognostic factor in non-Hodgkin’s lymphoma. Leuk. Lymphoma 2003, 44, 1733–1738. [Google Scholar]
- Yin, B.W.; Dnistrian, A.; Lloyd, K.O. Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int. J. Cancer 2002, 98, 737–740. [Google Scholar]
- Albert, L.J.; Inman, R.D. Molecular mimicry and autoimmunity. N. Engl. J. Med 1999, 341, 2068–2074. [Google Scholar]
- Rose, N.R.; Mackay, I.R. Molecular mimicry: A critical look at exemplary instances in human diseases. Cell Mol. Life Sci 2000, 57, 542–551. [Google Scholar]
- Welsh, R.M.; Fujinami, R.S. Pathogenic epitopes, heterologous immunity and vaccine design. Nat. Rev. Microbiol 2007, 5, 555–563. [Google Scholar]
- Schneider, J.; Moragues, D.; MartÃnez, N.; Romero, H.; Jimenez, E.; Pontin, J. Cross-Reactivity between Candida albicans and human ovarian carcinoma as revealed by monoclonal antibodies PA10F and C6. Br. J. Cancer 1998, 77, 1015–1020. [Google Scholar]
- Krause, I.; Blank, M.; Cervera, R.; Font, J.; Matthias, T.; Pfeiffer, S.; Wies, I.; Fraser, A.; Shoenfeld, Y. Cross-Reactive epitopes on beta2-glycoprotein-I and Saccharomyces cerevisiae in patients with the antiphospholipid syndrome. Ann. N. Y. Acad. Sci 2007, 1108, 481–488. [Google Scholar]
- Oshitani, N.; Hato, F.; Suzuki, K.; Sawa, Y.; Matsumoto, T.; Maeda, K.; Higuchi, K.; Kitagawa, S.; Arakawa, T. Cross-Reactivity of yeast antigens in human colon and peripheral leukocytes. J. Pathol 2003, 199, 361–367. [Google Scholar]
- Srinivasappa, J.; Saegusa, J.; Prabhakar, B.S.; Gentry, M.K.; Buchmeier, M.J.; Wiktor, T.J.; Koprowski, H.; Oldstone, M.B.; Notkins, A.L. Molecular mimicry: Frequency of reactivity of monoclonal antiviral antibodies with normal tissues. J. Virol 1986, 57, 397–401. [Google Scholar]
- Punta, M.; Ofran, Y. The rough guide to in silico function prediction, or how to use sequence and structure information to predict protein function. PLoS Comput. Biol 2008, 4, e1000160. [Google Scholar]
- Sleator, R.D.; Walsh, P. An overview of in silico protein function prediction. Arch. Microbiol 2010, 192, 151–155. [Google Scholar]
- Glucan 1,4-alpha-glucosidase activity. Available online: http://amigo.geneontology.org/cgi-bin/amigo/term-details.cgi?term=GO:0004339 accessed on 9 April 2012.
- GO:0008061 chitin binding. Available online: http://www.ebi.ac.uk/QuickGO/GTerm?id=GO:0008061 accessed on 9 April 2012.
- Cation transport. Available online: http://amigo.geneontology.org/cgi-bin/amigo/term-details.cgi?term=GO:0006812 accessed on 9 April 2012.
- ATP synthesis coupled proton transport. Available online: http://amigo.geneontology.org/cgi-bin/amigo/term-details.cgi?term=GO:0015986 accessed on 9 April 2012.
- Seelenmeyer, C.; Wegehingel, S.; Lechner, J.; Nickel, W. The cancer antigen CA125 represents a novel counter receptor for galectin-1. J. Cell Sci 2003, 116, 1305–1318. [Google Scholar]
- Rump, A.; Morikawa, Y.; Tanaka, M.; Minam, S.; Umesaki, N.; Takeuchi, M.; Miyajima, A. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J. Biol. Chem 2004, 279, 9190–9198. [Google Scholar]
- Patankar, M.S.; Jing, Y.; Morrison, J.C.; Belisle, J.A.; Lattanzio, F.A.; Deng, Y.; Wong, N.K.; Morris, H.R.; Dell, A.; Clark, G.F. Potent suppression of natural killer cell response mediated by the ovarian tumor marker. Gynecol. Oncol 2005, 99, 704–713. [Google Scholar]
- Gipson, I.K.; Blalok, T.; Tisdale, A.; Spurr-Michaud, S.; Allcorn, S.; Stavreus-Evers, A.; Gemzell, K. Muc16 is lost from the uterodome (pinopode) surface of the receptive human endometrium: In vitro evidence that MUC16 is a barrier to trophoblast adherence. Biol. Reprod 2008, 78, 134–142. [Google Scholar]
- Özkucur, N.; Perike, S.; Sharma, P.; Funk, R.H.W. Persistent directional cell migration requires ion transport proteins as direction sensors and membrane potential differences in order to maintain directedness. BMC Cell Biol 2011, 12. [Google Scholar] [CrossRef]
- Gloor, S.; Antonicek, H.; Sweadner, K.J.; Pagliusi, S.; Frank, R.; Moos, M.; Schachner, M. The adhesion molecule on glia (AMOG) is a homologue of the beta subunit of the Na,K-ATPase. J. Cell Biol 1990, 110, 165–174. [Google Scholar]
- Straver, M.H.; Smit, G.; Kijne, J.W. Purification and partial characterization of a flocculin from brewer’s yeast. Appl. Environ. Microbiol 1994, 60, 2754–2758. [Google Scholar]
- Adams, J.C. Cell-Matrix contact structures. Cell Mol. Life Sci 2001, 58, 371–392. [Google Scholar]
- Duraisamy, S.; Ramasamy, S.; Kharbanda, S.; Kufe, D. Distinct evolution of the human carcinoma-associated transmembrane mucins, MUC1, MUC4 and MUC16. Gene 2006, 373, 28–34. [Google Scholar]
- UniProtKB. Available online: http://www.uniprot.org/ accessed on 2 April 2012.
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov accessed on 30 March 2012.
- The UniProt Consortium (UniProt). Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012, 40, D71–D75.
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic local alignment search tool. J. Mol. Biol 1990, 215, 403–410. [Google Scholar]
- Mountm, D.W. Using the Basic amd Local Alignment Search Tool (BLAST). In Cold Spring Harbor Protocols; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2007; Volume 14, p. 17. [Google Scholar]
- Friedberg, I.; Harder, T.; Godzik, A. JAFA: A protein function annotation meta-server. Nucleic Acids Res 2006, 34, W379–W381. [Google Scholar]
- Joined Assembly of Function Annotations-JAFA. Available online: http://jafa.burnham.org/ accessed on 6 December 2006.
- Hawkins, T.; Luban, S.; Kihara, D. Enhanced automated function prediction using distantly related sequences and contextual association by PFP. Protein Sci 2006, 15, 1550–1556. [Google Scholar]
- Kihara Bioinformatics Laboratory. Available online: http://dragon.bio.purdue.edu/pfp accessed on 21 August 2008.
- Juhl Jensen, L.; Stærfeldt, H.H.; Brunak, S. Prediction of human protein function according to Gene Ontology categories. Bioinformatics 2003, 19, 635–642. [Google Scholar]
- GO Annotation Tools. Available online: http://www.geneontology.org/GO.tools.annotation.shtml accessed on 21 February 2012.
- Tatebayashi, K.; Tanaka, K.; Yang, H.Y.; Yamamoto, K.; Matsushita, Y.; Tomida, T.; Imai, M.; Saito, H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J 2007, 26, 3521–3533. [Google Scholar]
- Pelaseyed, T.; Hansson, G.C. CFTR anion channel modulates expression of human transmembrane mucin MUC3 through the PDZ protein GOPC. J. Cell Sci 2011, 124, 3074–3083. [Google Scholar]
- Cullen, P.J.; Spraguem, G.F. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell 2002, 13, 2990–3004. [Google Scholar]
- Bozzuto, G.; Ruggieri, P.; Molinari, A. Molecular aspects of tumor cell migration and invasion. Ann. Ist. Super. Sanita 2010, 46, 66–80. [Google Scholar]
| Database | Hit * | Protein/Conserved Domain ** | Organism | Length | Identity | Score | E-value |
|---|---|---|---|---|---|---|---|
| Virus | O39781/Q6SV6WO | MembraneGlycoprotein, gp2/Gp2 ** | Equine herpesvirus 1 | 806 | 25% | 295 | 1 × 10−23 |
| E2GKY4 | Glycoproteingp350-220/BLLF1 ** | Epstein Barr virus | 877 | 26% | 170 | 3 × 10−9 | |
| Bacteria | B2ISC7/Q97P71 | Cell surface anchor family protein | Streptococcus pneumoniae(strain GSP14) | 4695 | 17% | 669 | 4 × 10−66 |
| Q4L9PO | Serine-rich adhesion for platelets | Staphylococcus haemolyticus (strain JCSC1435) | 3608 | 20% | 625 | 5 × 10−61 | |
| Fungi | E9P8M0 | Cell surface flocculin | Saccharomyces cerevisiae | 1630 | 24% | 499 | 4 × 10−47 |
| C8ZAR8 | Muc1p | Saccharomyces cerevisiae | 1576 | 23% | 429 | 6 × 10−39 | |
| Eukaryota | E9AEM9 | Proteophosphoglycan 5 | Leishmania major | 17392 | 18% | 1003 | 6 × 10−105 |
| Biological process | Molecular function | Cellular component | ||||||
|---|---|---|---|---|---|---|---|---|
| GO | Score | Definition | GO | Score | Definition | GO | Score | Definition |
| 0006812 | 5363 | Cation transport | 0005515 | 3567 | Protein binding | 0005624 | 2,4775 | Membrane |
| 0007155 | 4679 | Cell adhesion | 0004867 | 3371 | Endopeptidase inhibitor activity | 0005887 | 4,420 | Integral to plasma membrane |
| 0007275 | 4273 | Development | 0004872 | 3133 | Receptor activity | 0016020 | 4,305 | Membrane |
| 0006929 | 3310 | Substrate-bound cell migration | 0004672 | 2914 | Protein kinase activity | 0005622 | 3,129 | Intracellular |
| 0050652 | 2664 | Polysachharide biosynthesis | 0004674 | 2623 | Ser/Thr kinase activity | 0016021 | 2,363 | Integral to membrane |
| 0007166 | 2554 | Cell surface receptor linked signal transduction | 0005529 | 2445 | Sugar binding | 0005578 | 2,118 | Extracellular matrix (sensu Metazoa) |
| 0007165 | 2416 | Signal transduction | 0005524 | 2219 | ATP binding | 0005634 | 1,939 | Nucleus |
| 0006917 | 2213 | Induction of apoptosis | 0008270 | 2206 | Zinc ion binding | 0005856 | 1,764 | Cytoskeleton |
| 0008228 | 2079 | Opsonization | 0003804 | 2154 | Coagulation factor Xa activity | 0009897 | 1,633 | External side of plasma membrane |
| 0035162 | 2071 | Embryonic hemopoiesis | 0046703 | 2075 | NK cell like-receptor binding | 0005615 | 1,531 | Extracellular space |
| Biological process | Molecular function | Cellular component | ||||||
|---|---|---|---|---|---|---|---|---|
| GO/GO-Level | Score | Name | GO/GO-Level | Score | Name | GO/GO-Level | Score | Name |
| 0015986/6 | 2.00 | ATP synthesis coupled proton transport | 0004339/6 | 2.00 | Glucan 1,4-alpha glucosidase activity | 0005886/3 | 1.00 | Plasma membrane |
| 0006030/6 | 2.00 | Chitin metabolism | 0005524/5 | 1.67 | ATP binding | 0016469/2 | 0.67 | Proton-transporting two-sector ATPase complex |
| 0000272/6 | 2.00 | Polysaccharide catabolism | 0008061/4 | 1.33 | Chitin binding | 0005615/2 | 0.67 | Extracellular space |
| 0007160/4 | 1.33 | Cell matrix adhesion | 0005515/2 | 0.67 | Protein binding | - | - | - |
| 0007124/3 | 1.00 | Pseudohyphal growth | 0005554/1 | 0.33 | Molecular function unknown | - | - | - |
| 0001403/3 | 1.00 | Invasive growth (sensu Saccharomyces) | - | - | - | - | - | - |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jankovic, M.; Mitic, N. MUC16/CA125 in the Context of Modular Proteins with an Annotated Role in Adhesion-Related Processes: In Silico Analysis. Int. J. Mol. Sci. 2012, 13, 10387-10400. https://doi.org/10.3390/ijms130810387
Jankovic M, Mitic N. MUC16/CA125 in the Context of Modular Proteins with an Annotated Role in Adhesion-Related Processes: In Silico Analysis. International Journal of Molecular Sciences. 2012; 13(8):10387-10400. https://doi.org/10.3390/ijms130810387
Chicago/Turabian StyleJankovic, Miroslava, and Ninoslav Mitic. 2012. "MUC16/CA125 in the Context of Modular Proteins with an Annotated Role in Adhesion-Related Processes: In Silico Analysis" International Journal of Molecular Sciences 13, no. 8: 10387-10400. https://doi.org/10.3390/ijms130810387




