Structural and Phylogenetic Analysis of Rhodobacter capsulatus NifF: Uncovering General Features of Nitrogen-fixation (nif)-Flavodoxins
Abstract
:1. Introduction
2. Results and Discussion
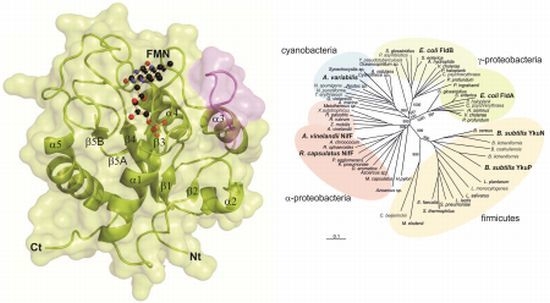
2.1. General Structure of Rc-NifF and FMN Environment
2.2. Distribution of Charged Residues
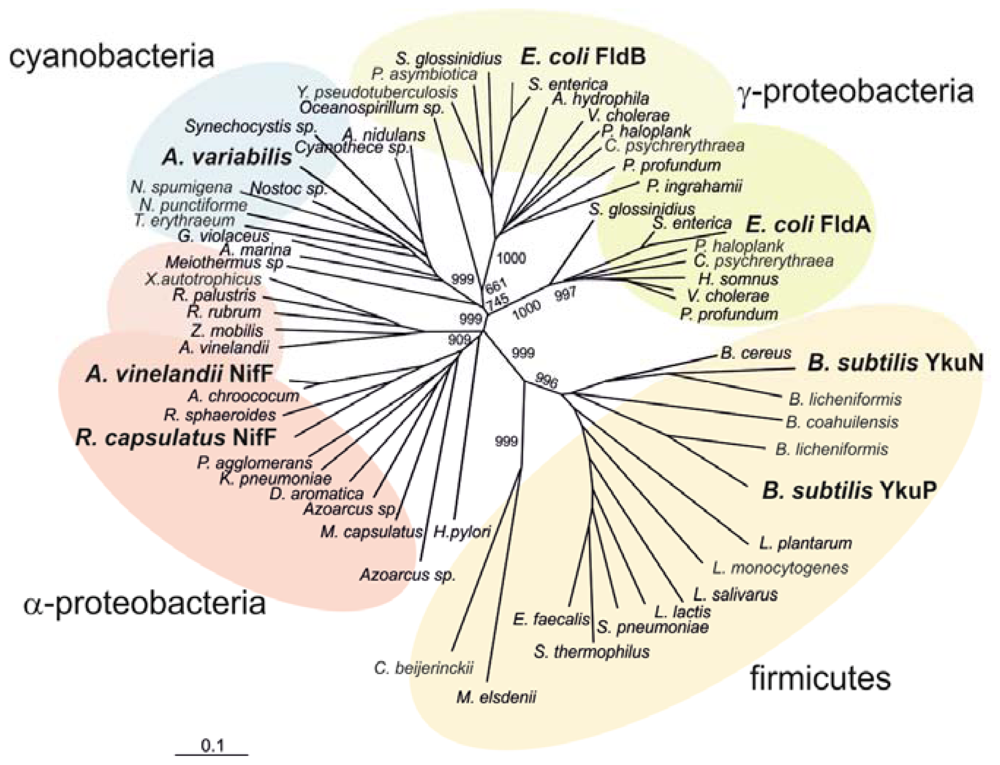
2.3. Phylogenetic Analysis
3. Experimental Section
3.1. Protein Expression and Purification
3.2. Crystallization and Data Collection
3.3. Structural Determination and Refinement
3.4. Phylogenetic Relationships
4. Conclusions
Acknowledgments
References
- Mayhew, S.G.; Tollin, G. General Properties of Flavodoxins. In Chemistry and Biochemistry of Flavoenzymes; Müller, F., Ed.; CRC Press: Boca Raton, FL, USA, 1993; pp. 389–426. [Google Scholar]
- Nogues, I.; Perez-Dorado, I.; Frago, S.; Bittel, C.; Mayhew, S.G.; Gomez-Moreno, C.; Hermoso, J.A.; Medina, M.; Cortez, N.; Carrillo, N. The ferredoxin-NADP(H) reductase from Rhodobacter capsulatus: Molecular structure and catalytic mechanism. Biochemistry 2005, 44, 11730–11740. [Google Scholar]
- Sancho, J. Flavodoxins: Sequence, folding, binding, function and beyond. Cell. Mol. Life Sci 2006, 63, 855–864. [Google Scholar]
- Lodeyro, A.F.; Ceccoli, R.D.; Pierella-Karlusich, J.J.; Carrillo, N. The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett 2012, 586, 2917–2924. [Google Scholar]
- Hallenbeck, P.C.; Gennaro, G. Stopped-flow kinetic studies of low potential electron carriers of the photosynthetic bacterium, Rhodobacter capsulatus: Ferredoxin I and NifF. Biochim. Biophys. Acta 1998, 1365, 435–442. [Google Scholar]
- Gennaro, G.; Hubner, P.; Sandmeier, U.; Yakunin, A.F.; Hallenbeck, P.C. Cloning, characterization, and regulation of nifF from Rhodobacter capsulatus. J. Bacteriol 1996, 178, 3949–3952. [Google Scholar]
- Yakunin, A.F.; Gennaro, G.; Hallenbeck, P.C. Purification and properties of a nif-specific flavodoxin from the photosynthetic bacterium Rhodobacter capsulatus. J. Bacteriol 1993, 175, 6775–6780. [Google Scholar]
- Hill, S.; Kavanagh, E.P. Roles of nifF and nifJ gene products in electron transport to nitrogenase in Klebsiella pneumoniae. J. Bacteriol 1980, 141, 470–475. [Google Scholar]
- Nieva-Gomez, D.; Roberts, G.P.; Klevickis, S.; Brill, W.J. Electron transport to nitrogenase in Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 1980, 77, 2555–2558. [Google Scholar]
- Klugkist, J.; Voorberg, J.J.; Haaker, H.; Veeger, C. Characterization of three different flavodoxins from Azotobacter vinelandii. Eur. J. Biochem 1986, 155, 33–40. [Google Scholar]
- Lowery, T.J.; Wilson, P.E.; Zhang, B.; Bunker, J.; Harrison, R.G.; Nyborg, A.C.; Thiriot, D.; Wat, G.D. Flavodoxin hydroquinone reduces Azotobacter vinelandii Fe protein to the all-ferrous redox state with a S = 0 spin state. Proc. Natl. Acad. Sci. USA 2006, 103, 17131–17136. [Google Scholar]
- Bittel, C.; Tabares, L.C.; Armesto, M.; Carrillo, N.; Cortez, N. The oxidant-responsive diaphorase of Rhodobacter capsulatus is a ferredoxin (flavodoxin)-NADP(H) reductase. FEBS Lett 2003, 553, 408–412. [Google Scholar]
- Alagaratnam, S.; van Pouderoyen, G.; Pijning, T.; Dijkstra, B.W.; Cavazzini, D.; Rossi, G.L.; van Dongen, W.M.; van Mierlo, C.P.; van Berkel, W.J.; Canters, G.W. A crystallographic study of Cys69Ala flavodoxin II from Azotobacter vinelandii: Structural determinants of redox potential. Protein Sci 2005, 14, 2284–2295. [Google Scholar]
- Lostao, A.; Gómez-Moreno, C.; Mayhew, S.G.; Sancho, J. Differential stabilization of the three FMN redox forms by tyrosine 94 and tryptophan 57 in flavodoxin from Anabaena and its influence on the redox potentials. Biochemistry 1997, 36, 14334–14344. [Google Scholar]
- Drummond, M.H. Structure predictions and surface charge of nitrogenase flavodoxins from Klebsiella pneumoniae and Azotobacter vinelandii. Eur. J. Biochem 1986, 159, 549–553. [Google Scholar]
- Peelen, S.; Wijmenga, S.; Erbel, P.J.; Robson, R.L.; Eady, R.R.; Vervoort, J. Possible role of a short extra loop of the long-chain flavodoxin from Azotobacter chroococcum in electron transfer to nitrogenase: Complete 1H, 15N and 13C backbone assignments and secondary solution structure of the flavodoxin. J. Biomol. NMR 1996, 7, 315–330. [Google Scholar]
- Lawson, R.J.; von Wachenfeldt, C.; Haq, I.; Perkins, J.; Munro, A.W. Expression and characterization of the two flavodoxin proteins of Bacillus subtilis, YkuN and YkuP: Biophysical properties and interactions with cytochrome P450. Biol. Biochem 2004, 43, 12390–12409. [Google Scholar]
- Malca, S.H.; Girhard, M.; Schuster, S.; Durre, P.; Urlacher, V.B. Expression, purification and characterization of two Clostridium acetobutylicum flavodoxins: Potential electron transfer partners for CYP152A2. Biochim. Biophys. Acta 2011, 1814, 257–264. [Google Scholar]
- Geoghegan, S.M.; Mayhew, S.G.; Yalloway, G.N.; Butler, G. Cloning, sequencing and expression of the gene for flavodoxin from Megasphaera elsdenii and the effects of removing the protein negative charge that is closest to N(1) of the bound FMN. Eur. J. Biochem 2000, 267, 4434–4444. [Google Scholar]
- Gu, M.; Imlay, J.A. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol 2011, 79, 1136–1150. [Google Scholar]
- Giro, M.; Carrillo, N.; Krapp, A.R. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology 2006, 152, 1119–1128. [Google Scholar]
- Knappe, J.; Blaschkowski, H.P.; Grobner, P.; Schmitt, T. Pyruvate formate-lyase of Escherichia coli: The acetyl-enzyme intermediate. Eur. J. Biochem 1974, 50, 253–263. [Google Scholar]
- Bianchi, V.; Eliasson, R.; Fontecave, M.; Mulliez, E.; Hoover, D.M.; Matthews, R.G.; Reichard, P. Flavodoxin is required for the activation of the anaerobic ribonucleotide reductase. Biochem. Biophys. Res. Commun 1993, 197, 792–797. [Google Scholar]
- Gaudu, P.; Weiss, B. Flavodoxin mutants of Escherichia coli K-12. J. Bacteriol 2000, 182, 1788–1793. [Google Scholar]
- Bagby, S.; Barker, P.D.; Hill, H.A.; Sanghera, G.S.; Dunbar, B.; Ashby, G.A.; Eady, R.R.; Thorneley, R.N. Direct electrochemistry of two genetically distinct flavodoxins isolated from Azotobacter chroococcum grown under nitrogen-fixing conditions. Biochem. J 1991, 277, 313–319. [Google Scholar]
- Bennett, L.T.; Jacobson, M.R.; Dean, D.R. Isolation, sequencing, and mutagenesis of the nifF gene encoding flavodoxin from Azotobacter vinelandii. J. Biol. Chem 1988, 263, 1364–1369. [Google Scholar]
- Deistung, J.; Cannon, F.C.; Cannon, M.C.; Hill, S.; Thorneley, R.N. Electron transfer to nitrogenase in Klebsiella pneumoniae. nifF gene cloned and the gene product, a flavodoxin, purified. Biochem. J 1985, 231, 743–753. [Google Scholar]
- Medina, M. Structural and mechanistic aspects of flavoproteins: Photosynthetic electron transfer from photosystem I to NADP+. FEBS J 2009, 276, 3942–3958. [Google Scholar]
- Setif, P. Ferredoxin and flavodoxin reduction by photosystem I. Biochim. Biophys. Acta 2001, 1507, 161–179. [Google Scholar]
- Razquin, P.; Schmitz, S.; Peleato, M.L.; Fillat, M.F.; Gómez-Moreno, C.; Böhme, H. Differential activities of heterocyst ferredoxin, vegetative cell ferredoxin, and flavodoxin as electron carriers in nitrogen fixation and photosynthesis in Anabaena sp. Photosynthesis Res 1995, 43, 35–40. [Google Scholar]
- Masepohl, B.; Scholisch, K.; Gorlitz, K.; Kutzki, C.; Bohme, H. The heterocyst-specific fdxH gene product of the cyanobacterium Anabaena sp. PCC 7120 is important but not essential for nitrogen fixation. Mol. Gen. Genet 1997, 253, 770–776. [Google Scholar]
- Perez-Dorado, I.; Bortolotti, A.; Cortez, N.; Hermoso, J.A. Crystallization of a flavodoxin involved in nitrogen fixation in Rhodobacter capsulatus. Acta Crystallogr. Sect. F 2008, 64, 375–377. [Google Scholar]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D 2010, 66, 22–25. [Google Scholar]
- Brunger, A.T.; Adams, P.D.; Rice, L.M. Recent developments for the efficient crystallographic refinement of macromolecular structures. Curr. Opin. Struct. Biol 1998, 8, 606–611. [Google Scholar]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 1997, 53, 240–255. [Google Scholar]
- Jones, T..A.; Zou, J.Y.; Cowan, S.W.; Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 1991, 47, 110–119. [Google Scholar]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst 1993, 26, 283–291. [Google Scholar]
- Larkin, M.A.; Blackshields, M.A.G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol 1987, 4, 406–425. [Google Scholar]


| Data collection statistics | |
|---|---|
| Space group, unit cell (Å, °) | P41212, a = b = 66.49 c = 121.32 α = β = γ = 90 |
| Temperature (K) | 100 |
| Wavelength (Å) | 1.5418 |
| No. of molecules/a.u. | 1 |
| Resolution (Å) | 25.65–(2.35–2.17) |
| No. observations | 563236 |
| No. unique observations | 24830 |
| Redundancy | 22.4 (22.9) |
| Completeness (%) | 99.9 (100) |
| I/σ (I) | 26.0 (8.2) |
| Rsymb | 0.07 (0.49) |
| Refinement statistics | |
| Resolution range (Å) | 25.65–2.17 |
| Rwork | 0.25 |
| Rfreec | 0.27 |
| No. of non-hydrogen atoms | |
| Protein | 1402 |
| Ligand | 31 |
| Solvent | 76 |
| RMS deviations from ideal | |
| Rmsd bond length (Å) | 0.006 |
| Rmsd bond angles (°) | 1.4 |
| Ramachandran Plot | |
| Most favored (%) | 89.6 |
| Additionally allowed (%) | 9.7 |
| Generously allowed (%) | 0.6 |
| Average B-factor (Å2) | 43.5 |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pérez-Dorado, I.; Bortolotti, A.; Cortez, N.; Hermoso, J.A. Structural and Phylogenetic Analysis of Rhodobacter capsulatus NifF: Uncovering General Features of Nitrogen-fixation (nif)-Flavodoxins. Int. J. Mol. Sci. 2013, 14, 1152-1163. https://doi.org/10.3390/ijms14011152
Pérez-Dorado I, Bortolotti A, Cortez N, Hermoso JA. Structural and Phylogenetic Analysis of Rhodobacter capsulatus NifF: Uncovering General Features of Nitrogen-fixation (nif)-Flavodoxins. International Journal of Molecular Sciences. 2013; 14(1):1152-1163. https://doi.org/10.3390/ijms14011152
Chicago/Turabian StylePérez-Dorado, Inmaculada, Ana Bortolotti, Néstor Cortez, and Juan A. Hermoso. 2013. "Structural and Phylogenetic Analysis of Rhodobacter capsulatus NifF: Uncovering General Features of Nitrogen-fixation (nif)-Flavodoxins" International Journal of Molecular Sciences 14, no. 1: 1152-1163. https://doi.org/10.3390/ijms14011152