Cloning, Expression and Characterization of Mitochondrial Manganese Superoxide Dismutase from the Whitefly, Bemisia tabaci
Abstract
:1. Introduction
2. Results
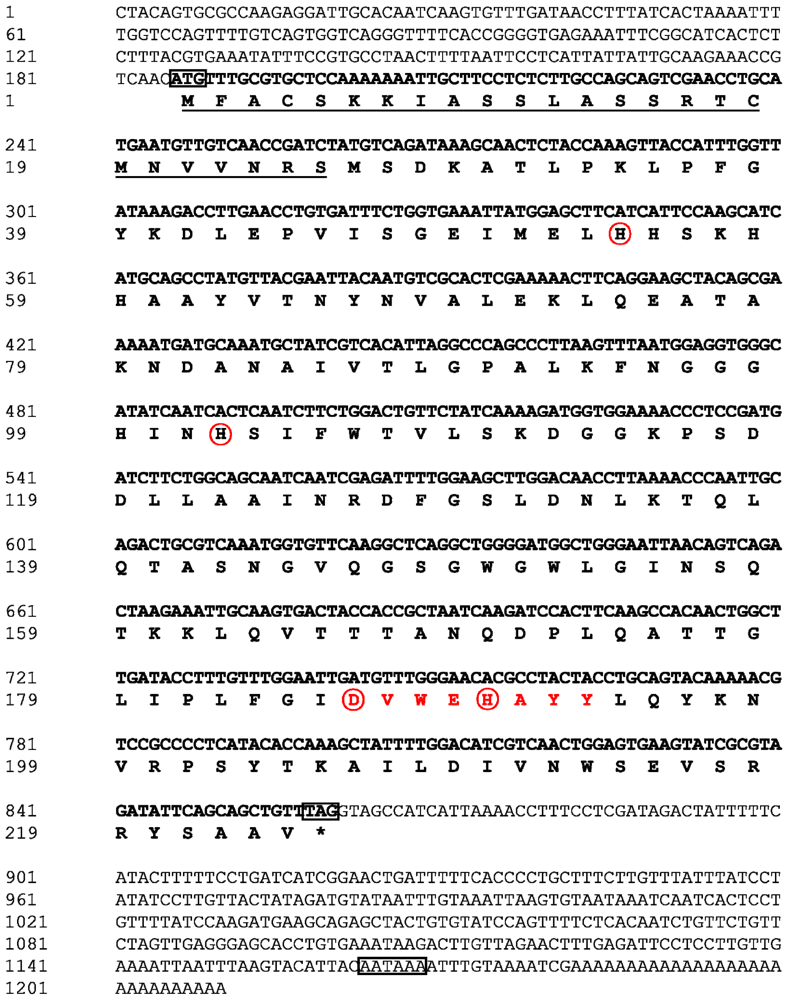
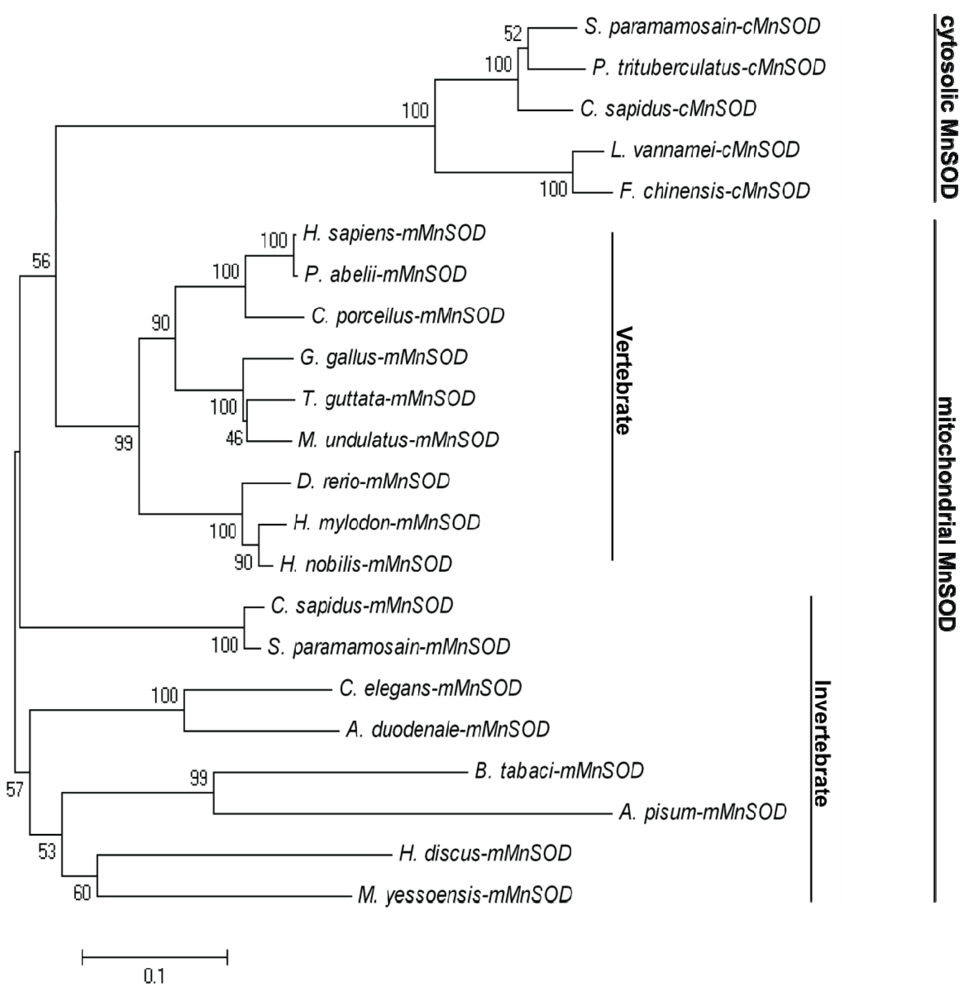
2.1. Analysis of the Bt-mMnSOD Sequence and the Predicted Protein
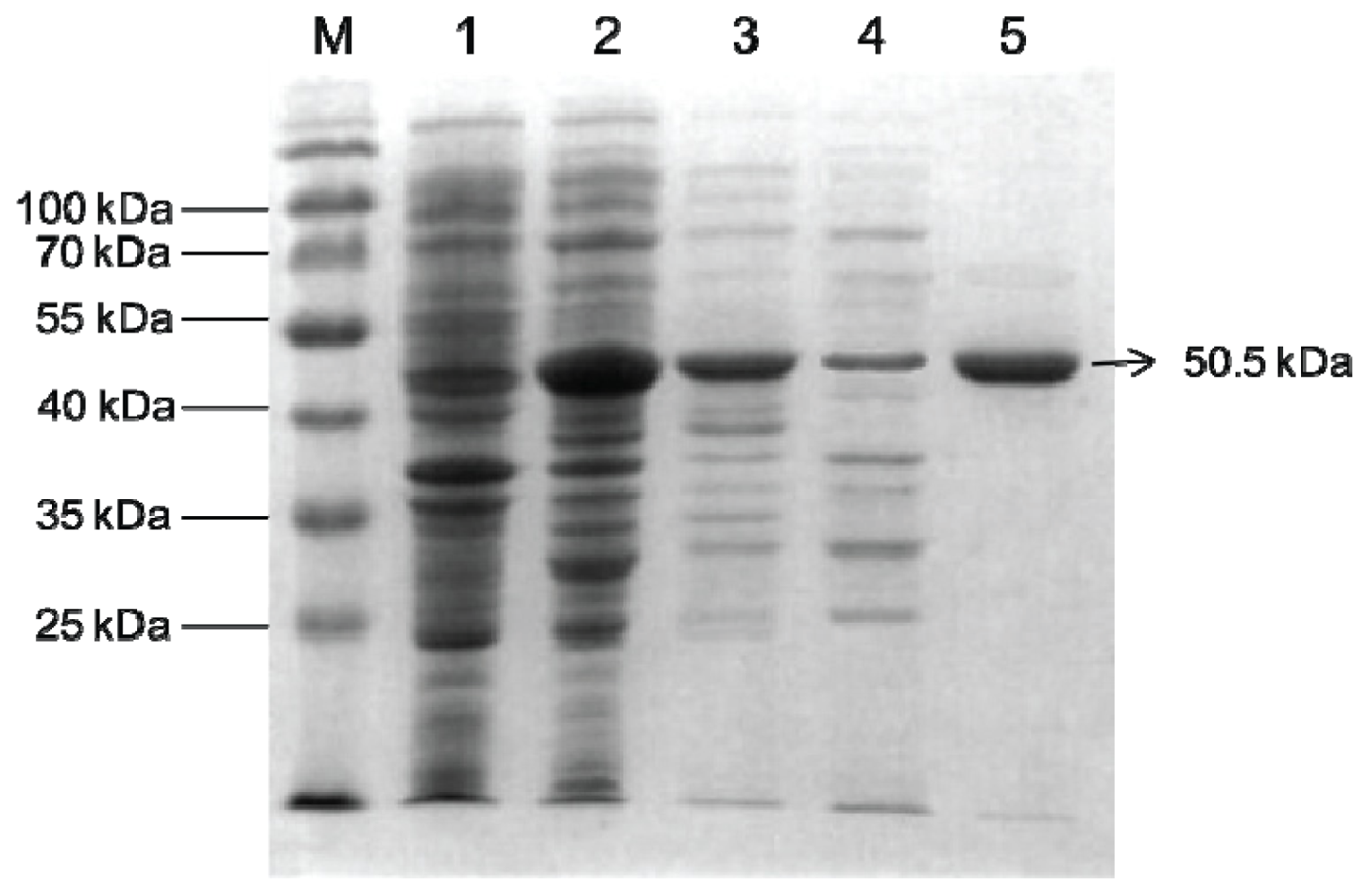
2.2. Overexpression and Purification of Bt-mMnSOD
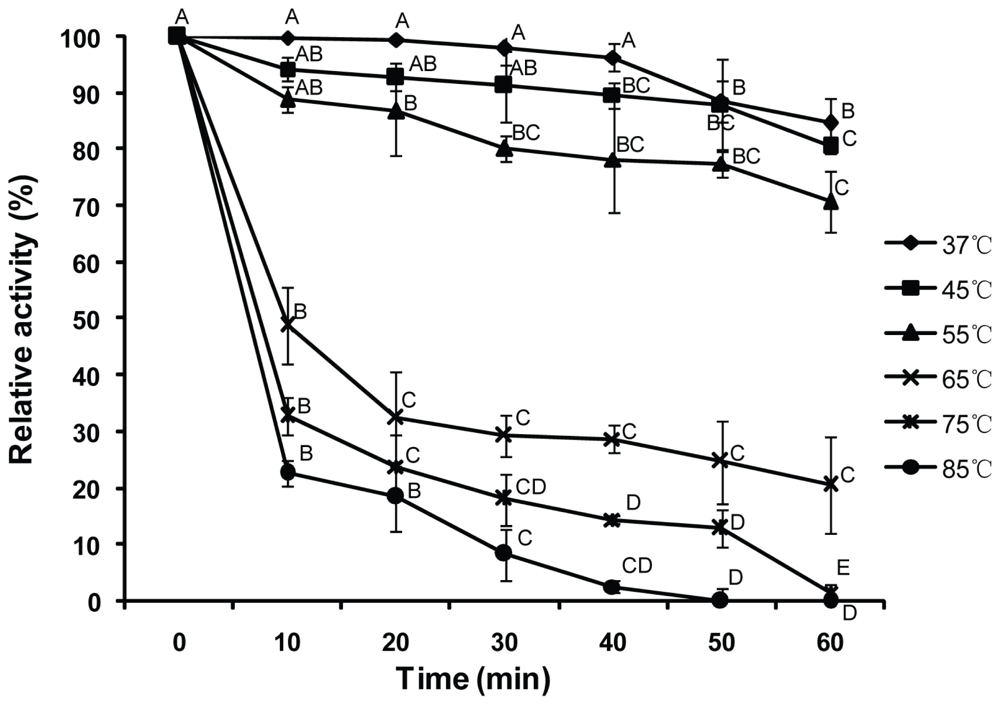
2.3. Thermostability of Purified Bt-mMnSOD
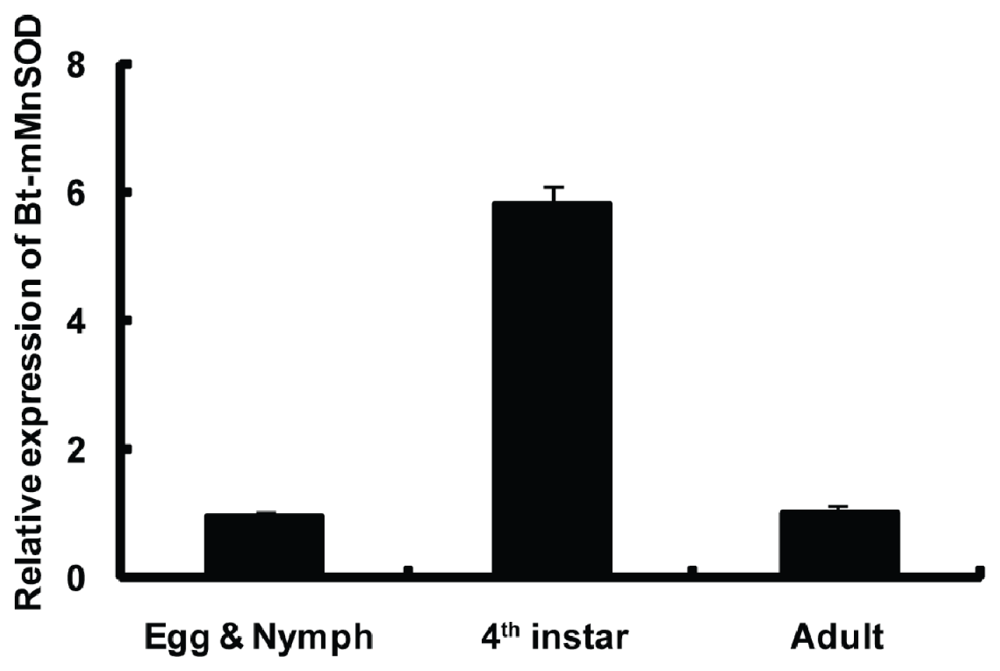
2.4. Expression of Bt-mMnSOD during Development
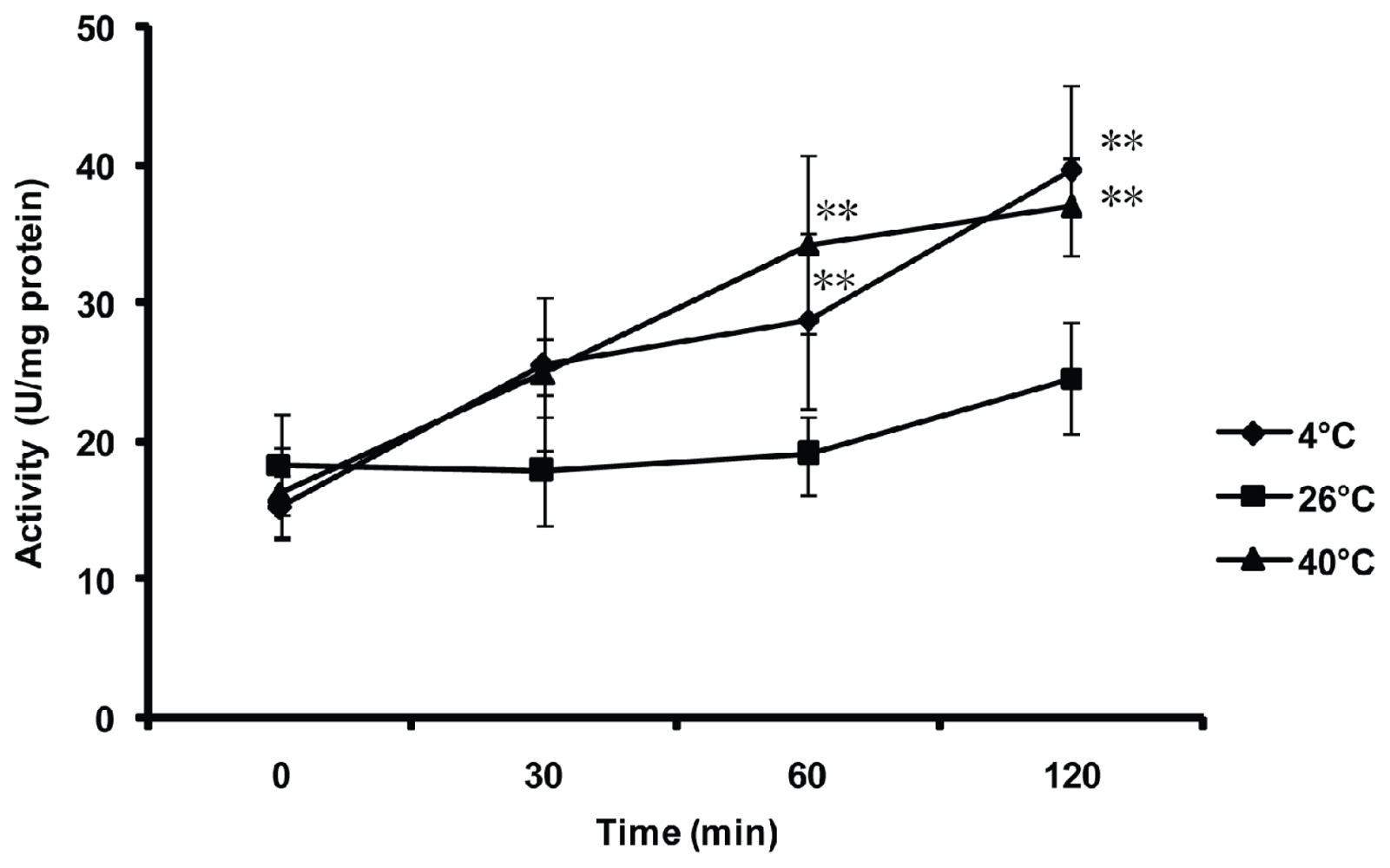
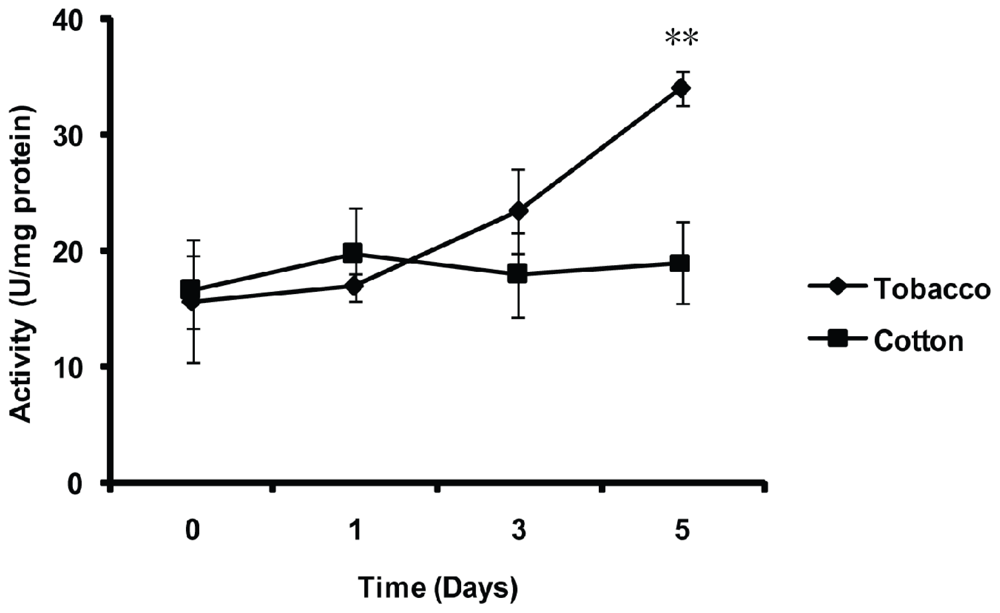
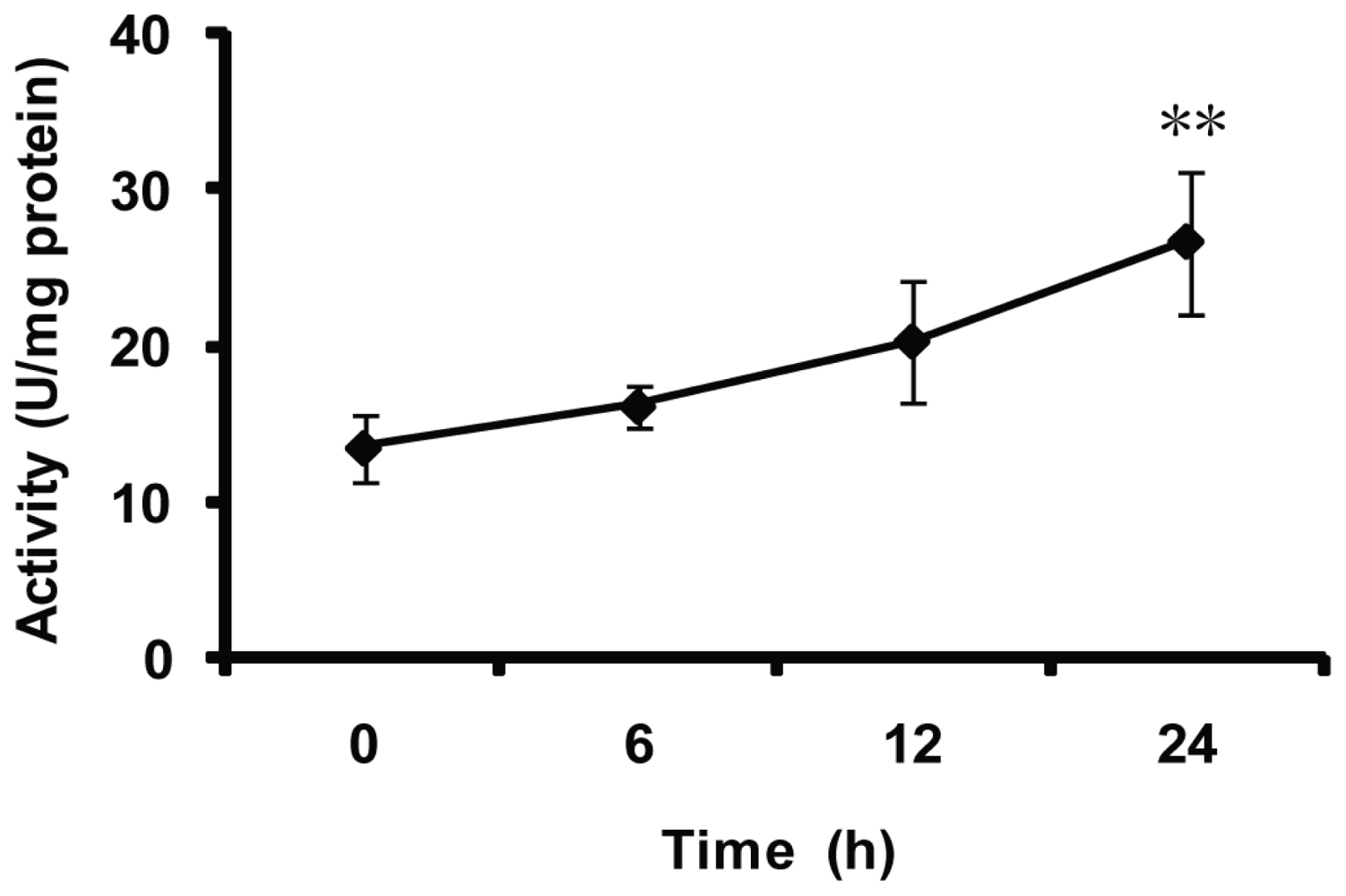
2.5. Effect of Thermal Stresses, Host Shift and Pesticide Treatment on MnSOD Activity
3. Discussion
4. Experimental Section
4.1. Whitefly Cultures and Plants
4.2. cDNA Synthesis and Cloning of the Bt-mMnSOD
4.3. Sequence Analysis
4.4. Overexpression and Purification of Bt-mMnSOD
4.5. Thermostability of Purified Bt-mMnSOD
4.6. Bt-mMnSOD Gene Expression in Different Developmental Stages
4.7. Effect of Thermal Stresses on the MnSOD Activity of the Whitefly
4.8. Effect of Host Shift on the MnSOD Activity of the Whitefly
4.9. Effect of Pesticide Treatment on MnSOD Activity of the Whitefly
5. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. The sweetpotato or silverleaf whiteflies: Biotypes of Bemisia tabaci or a species complex? Annu. Rev. Entomol 1995, 40, 511–534. [Google Scholar]
- Perring, T.M. The Bemisia tabaci species complex. Crop Prot 2001, 20, 725–737. [Google Scholar]
- Boykin, L.M.; Shatters, R.G., Jr; Rosell, R.C.; McKenzie, C.L.; Bagnall, R.A.; de Barro, P.J.; Frohlich, D.R. Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol. Phylogenet. Evol. 2007, 44, 1306–1319. [Google Scholar]
- De Barro, P.J.; Liu, S.S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol 2011, 56, 1–19. [Google Scholar]
- De Barro, P.J. Bemisia tabaci, a top 100 invader. J. Insect Sci 2008, 8, 16. [Google Scholar]
- Dalton, R. Whitefly infestations: The Christmas invasion. Nature 2006, 443, 898–900. [Google Scholar]
- Liu, S.S.; de Barro, P.; Xu, J.; Luan, J.B.; Zang, L.S.; Ruan, Y.M.; Wan, F.H. Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 2007, 318, 1769–1772. [Google Scholar]
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; de Barro, P.J. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am 2010, 103, 196–208. [Google Scholar]
- Luo, C.; Yao, Y.; Wang, R.J.; Yan, F.M.; Hu, D.X.; Zhang, Z.L. The use of mitochondrial cytochrome oxidase I (mt COI) gene sequences for the identification of biotypes of Bemisia tabaci (Gennadius) in China. Acta Entomol. Sin 2002, 45, 759–763. [Google Scholar]
- Zhang, L.P.; Zhang, Y.J.; Zhang, W.J.; Wu, Q.J.; Xu, B.Y.; Chu, D. Analysis of genetic diversity among different geographical populations and determination of biotypes of Bemisia tabaci in China. J. Appl. Entomol 2005, 129, 121–128. [Google Scholar]
- Chu, D.; Zhang, Y.J.; Brown, J.K.; Cong, B.; Xu, B.Y.; Wu, Q.J.; Zhu, G.R. The introduction of the exotic Q biotype of Bemisia tabaci from the Mediterranean region into China on ornamental crops. Fla. Entomol 2006, 89, 168–174. [Google Scholar]
- Xu, J. Studies on the Invasion by Alien Bemisia tabaci in Zhejiang and Comparison of Biological Characteristics between Biotypes of the Whitefly. Ph.D. Thesis, Zhejiang University, Hangzhou, China, June 2009. [Google Scholar]
- Oliveira, M.R.V.; Henneberry, T.J.; Anderson, P. History, current status and collaborative research projects for Bemisia tabaci. Crop Prot 2001, 20, 709–723. [Google Scholar]
- Rosell, R.C.; Shah, A.; Khan, M.; Aghakasiri, N.; Shatters, R.G.; McKenzie, C.L. Superoxide dismutase activity in temperature stressed viruliferous whiteflies (Hemiptera: Aleyrodidae). Preceeding of the XXIII International Congress of Entomology, Durban, South Africa, 6–12 July 2008.
- Li, J.M.; Su, Y.L.; Gao, X.L.; He, J.; Liu, S.S.; Wang, X.W. Molecular characterization and oxidative stress response of an intracellular Cu/Zn superoxide dismutase (CuZnSOD) of the whitefly, Bemisia tabaci. Arch. Insect Biochem 2011, 77, 118–133. [Google Scholar]
- McCord, J.M.; Fridovich, I. The utility of superoxide dismutase in studying free radical reactions. J. Biol. Chem 1969, 244, 6056–6063. [Google Scholar]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Mates, J.M.; Perez-Gomez, C.; de Castro, I.N. Antioxidant enzymes and human diseases. Clin. Biochem 1999, 32, 595–603. [Google Scholar]
- Brouwer, M.; Brouwer, T.H.; Grater, W.; Brown-Peterson, N. Replacement of a cytosolic copper/zinc superoxide dismutase by a novel cytosolic manganese superoxide dismutase in crustaceans that use copper (haemocyanin) for oxygen transport. Biochem. J 2003, 374, 219–228. [Google Scholar]
- Plantivaux, A.; Furla, P.; Zoccola, D.; Garello, G.; Forcioli, D.; Richier, S.; Merle, P.L.; Tambutté, É.; Tambutté, S.; Allemand, D. Molecular characterization of two CuZn-superoxide dismutases in a sea anemone. Free Radic. Biol. Med. 2004, 37, 1170–1181. [Google Scholar]
- Bannister, J.V.; Bannister, W.H.; Rotilio, G. Aspects of the structure, function and application of superoxide dismutase. CRC Crit. Rev. Biochem 1987, 22, 111–180. [Google Scholar]
- Ken, C.F.; Lee, C.C.; Duan, K.J.; Lin, C.T. Unusual stability of manganese superoxide dismutase from a new species, Tatumella ptyseos ct: Its gene structure, expression and enzyme properties. Protein Expres. Purif 2005, 40, 42–50. [Google Scholar]
- Brouwer, M.; Brouwer, T.H.; Grater, W.; Enghild, J.J.; Thogersen, I.B. The paradigm that all oxygen-respiring eukaryotes have cytosolic CuZn-superoxide dismutase and that Mn-superoxide dismutase is localized to the mitochondria does not apply to a large group of marine arthropods. Biochemistry 1997, 36, 13381–13388. [Google Scholar]
- Beyer, W.; Imlay, J.; Fridovich, I. Superoxide dismutases. Prog. Nucleic Acid Re 1991, 40, 221–253. [Google Scholar]
- Cheng, W.; Tung, Y.H.; Liu, C.H.; Chen, J.C. Molecular cloning and characterisation of cytosolic manganese superoxide dismutase (cytMn-SOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immun 2006, 20, 438–449. [Google Scholar]
- Lin, Y.C.; Lee, F.F.; Wu, C.L.; Chen, J.C. Molecular cloning and characterization of a cytosolic manganese superoxide dismutase (cytMnSOD) and mitochondrial manganese superoxide dismutase (mtMnSOD) from the kuruma shrimp Marsupenaeus japonicus. Fish Shellfish Immun 2010, 28, 143–150. [Google Scholar]
- Sun, J.; Folk, D.; Bradley, T.J.; Tower, J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics 2002, 161, 661–672. [Google Scholar]
- Greenberger, J.S.; Epperly, M.W.; Gretton, J.; Jefferson, M.; Nie, S.; Bernarding, M.; Kagan, V.; Guo, H.L. Radioprotective gene therapy. Curr. Gene Ther. 2003, 3, 183–195. [Google Scholar]
- Greenberger, J.S.; Epperly, M.W. Radioprotective antioxidant gene therapy: Potential mechanisms of action. Gene Ther. Mol. Biol 2004, 8, 31–44. [Google Scholar]
- Kim, Y.I.; Kim, H.J.; Kwon, Y.M.; Kang, Y.J.; Lee, I.H.; Jin, B.R.; Han, Y.S.; Cheon, H.M.; Ha, N.G.; Seo, S.J. Modulation of MnSOD protein in response to different experimental stimulation in Hyphantria cunea. Comp. Biochem. Phys. B 2010, 157, 343–350. [Google Scholar]
- Wang, M.Q.; Su, X.R.; Li, Y.; Jun, Z.; Li, T.W. Cloning and expression of the Mn-SOD gene from Phascolosoma esculenta. Fish Shellfish Immun 2010, 29, 759–764. [Google Scholar]
- Boveris, A.; Chance, B. Mitochondrial generation of hydrogen-peroxide-general properties and effect of hyperbaric-oxygen. Biochem. J 1973, 134, 707–716. [Google Scholar]
- Cadenas, E.; Davies, K.J. Mitochondrial free radical generation, oxidative stress and aging. Free Radic. Bio. Med 2000, 29, 222–230. [Google Scholar]
- Harari, P.M.; Fuller, D.J.; Gerner, E.W. Heat-shock stimulates polyamine oxidation by 2 distinct mechanisms in mammalian-cell cultures. Int. J. Radiat. Oncol 1989, 16, 451–457. [Google Scholar]
- Rauen, U.; Polzar, B.; Stephan, H.; Mannherz, H.G.; de Groot, H. Cold-induced apoptosis in cultured hepatocytes and liver endothelial cells: Mediation by reactive oxygen species. FASEB J 1999, 13, 155–168. [Google Scholar]
- Banerjee Mustafi, S.; Chakraborty, P.K.; Dey, R.S.; Raha, S. Heat stress upregulates chaperone heat shock protein 70 and antioxidant manganese superoxide dismutase through reactive oxygen species (ROS), p38MAPK and Akt. Cell Stress Chaperon 2009, 14, 579–589. [Google Scholar]
- Dash, B.; Metz, R.; Huebner, H.J.; Porter, W.; Phillips, T.D. Molecular characterization of two superoxide dismutases from Hydra vulgaris. Gene 2007, 387, 93–108. [Google Scholar]
- Futuyma, D.J.; Peterson, S.C. Genetic-variation in the use of resources by insects. Annu. Rev. Entomol 1985, 30, 217–238. [Google Scholar]
- Rossiter, M.C. Genetic and phenotypic variation in diet breadth in a generalist herbivore. Evol. Ecol 1987, 1, 272–282. [Google Scholar]
- Janković-Hladni, M.; Ivanović, J.; Spasić, M.; Blagojević, D.; Perić-Mataruga, V. Effect of the host plant on the antioxidative defence in the midgut of Lymantria dispar L. caterpillars of different population origins. J. Insect Physiol 1997, 43, 101–106. [Google Scholar]
- Zang, L.S.; Fu, R.X.; Liu, S.S.; Li, J.M.; Liu, Y.Q. Comparison of susceptibility to insecticides between the B biotype and a non B biotype of Bemisia tabaci in Zhejiang (In Chinese). Chin. Bull. Entomol 2006, 43, 207–210. [Google Scholar]
- Kemal, B. Malathion-induced oxidative stress in a parasitoid wasp: Effect on adult emergence, longevity, fecundity and oxidative and antioxidative response of Pimpla turionellae (Hymenoptera : Ichneumonidae). J. Econ. Entomol 2006, 99, 1225–1234. [Google Scholar]
- Jiu, M.; Zhou, X.P.; Tong, L.; Xu, J.; Yang, X.; Wan, F.H.; Liu, S.S. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS One 2007, 2, e182. [Google Scholar]
- DeBarro, P.J.; Driver, F. Use of RAPD PCR to distinguish the B biotype from other biotypes of Bemisia tabaci (Gennadius)(Hemiptera: Aleyrodidae). Aust. J. Entomol 1997, 36, 149–152. [Google Scholar]
- Zang, L.S.; Chen, W.Q.; Liu, S.S. Comparison of performance on different host plants between the B biotype and a non-B biotype of Bemisia tabaci from Zhejiang, China. Entomol. Exp. Appl 2006, 121, 221–227. [Google Scholar]
- Xu, J.; Lin, K.K.; Liu, S.S. Performance on different host plants of an alien and an indigenous Bemisia tabaci from China. J. Appl. Entomol 2011, 135, 771–779. [Google Scholar]
- Wang, X.W.; Luan, J.B.; Li, J.M.; Su, Y.L.; Xia, J.; Liu, S.S. Transcriptome analysis and comparison reveal divergence between two invasive whitefly cryptic species. BMC Genomics 2011, 12, 458. [Google Scholar]
- DNAMAN. Available online: http://www.lynnon.com accessed on 10 June 2012.
- BLAST Search Program. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 1 March 2012.
- Prot-Param Tools. Available online: http://kr.expasy.org/tools/protparam.html accessed on 1 June 2012.
- Signal P Program. Available online: http://www.cbs.dtu.dk/services/SignalP accessed on 25 May 2012.
- MITOPROT. Available online: http://ihg.gsf.de/ihg/mitoprot.html accessed on 10 June 2012.
- TargetP. Available online: http://www.cbs.dtu.dk/services/TargetP accessed on 10 June 2012.
- Clustal W. Available online: http://www.ebi.ac.uk/Tools/msa/clustalw2 accessed on 1 May 2012.
- MEGA. Available online: http://www.megasoftware.net accessed on 1 April 2012.
- Guan, K.L.; Dixon, J.E. Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem 1991, 192, 262. [Google Scholar]
- Tang, Q.Y.; Feng, M.G. DPS Data Processing System; Science Press: Beijing, China, 2007. [Google Scholar]
- GenScript Real-time PCR Primer Design Software. Available online: https://www.genscript.com/ssl-bin/app/primer accessed on 27 June, 2012.

| Species | Protein | Abbreviation | GenBank no. |
|---|---|---|---|
| Homo sapiens | mitochondrial MnSOD | H. sapiens-mMnSOD | P04179.2 |
| Cavia porcellus | mitochondrial MnSOD | C. porcellus-mMnSOD | XM_003466367.1 |
| Pongo abelii | mitochondrial MnSOD | P. abelii-mMnSOD | NM_001133563.1 |
| Taeniopygia guttata | mitochondrial MnSOD | T. guttata-mMnSOD | DQ214967.1 |
| Gallus gallus | mitochondrial MnSOD | G. gallus-mMnSOD | NM_204211.1 |
| Melopsittacus undulatus | mitochondrial MnSOD | M. undulatus-mMnSOD | AY241394.1 |
| Danio rerio | mitochondrial MnSOD | D. rerio-mMnSOD | NP_956270.1 |
| Hemibarbus mylodon | mitochondrial MnSOD | H. mylodon-mMnSOD | ACR23311.1 |
| Hypophthalmichthys nobilis | mitochondrial MnSOD | H. nobilis-mMnSOD | ADM26563.1 |
| Bemisia tabaci | mitochondrial MnSOD | B. tabaci-mMnSOD | JQ867105 |
| Acyrthosiphon pisum | mitochondrial MnSOD | A. pisum-mMnSOD | NM_001246048.1 |
| Caenorhabditis elegans | mitochondrial MnSOD | C. elegans-mMnSOD | D12984.1 |
| Ancylostoma duodenale | mitochondrial MnSOD | A. duodenale-mMnSOD | FJ465146.1 |
| Haliotis discus discus | mitochondrial MnSOD | H. discus-mMnSOD | DQ821491.1 |
| Mizuhopecten yessoensis | mitochondrial MnSOD | M. yessoensis-mMnSOD | AB222783.1 |
| Callinectes sapidus | mitochondrial MnSOD | C. sapidus-mMnSOD | AF264029.1 |
| cytosolic MnSOD | C. sapidus-cMnSOD | AAF74771.1 | |
| Scylla paramamosain | mitochondrial MnSOD | S. paramamosain-mMnSOD | FJ605170.2 |
| cytosolic MnSOD | S. paramamosain-cMnSOD | ADA63848.1 | |
| Portunus trituberculatus | cytosolic MnSOD | P. trituberculatus-cMnSOD | FJ031018.1 |
| Fenneropenaeus chinensis | cytosolic MnSOD | F. chinensis-cMnSOD | ACS49842.1 |
| Litopenaeus vannamei | cytosolic MnSOD | L. vannamei-cMnSOD | DQ005531.1 |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gao, X.-L.; Li, J.-M.; Wang, Y.-L.; Jiu, M.; Yan, G.-H.; Liu, S.-S.; Wang, X.-W. Cloning, Expression and Characterization of Mitochondrial Manganese Superoxide Dismutase from the Whitefly, Bemisia tabaci. Int. J. Mol. Sci. 2013, 14, 871-887. https://doi.org/10.3390/ijms14010871
Gao X-L, Li J-M, Wang Y-L, Jiu M, Yan G-H, Liu S-S, Wang X-W. Cloning, Expression and Characterization of Mitochondrial Manganese Superoxide Dismutase from the Whitefly, Bemisia tabaci. International Journal of Molecular Sciences. 2013; 14(1):871-887. https://doi.org/10.3390/ijms14010871
Chicago/Turabian StyleGao, Xian-Long, Jun-Min Li, Yong-Liang Wang, Min Jiu, Gen-Hong Yan, Shu-Sheng Liu, and Xiao-Wei Wang. 2013. "Cloning, Expression and Characterization of Mitochondrial Manganese Superoxide Dismutase from the Whitefly, Bemisia tabaci" International Journal of Molecular Sciences 14, no. 1: 871-887. https://doi.org/10.3390/ijms14010871




