Investigations of the Binding of [Pt2(DTBPA)Cl2](II) and [Pt2(TPXA)Cl2](II) to DNA via Various Cross-Linking Modes
Abstract
:1. Introduction
2. Results
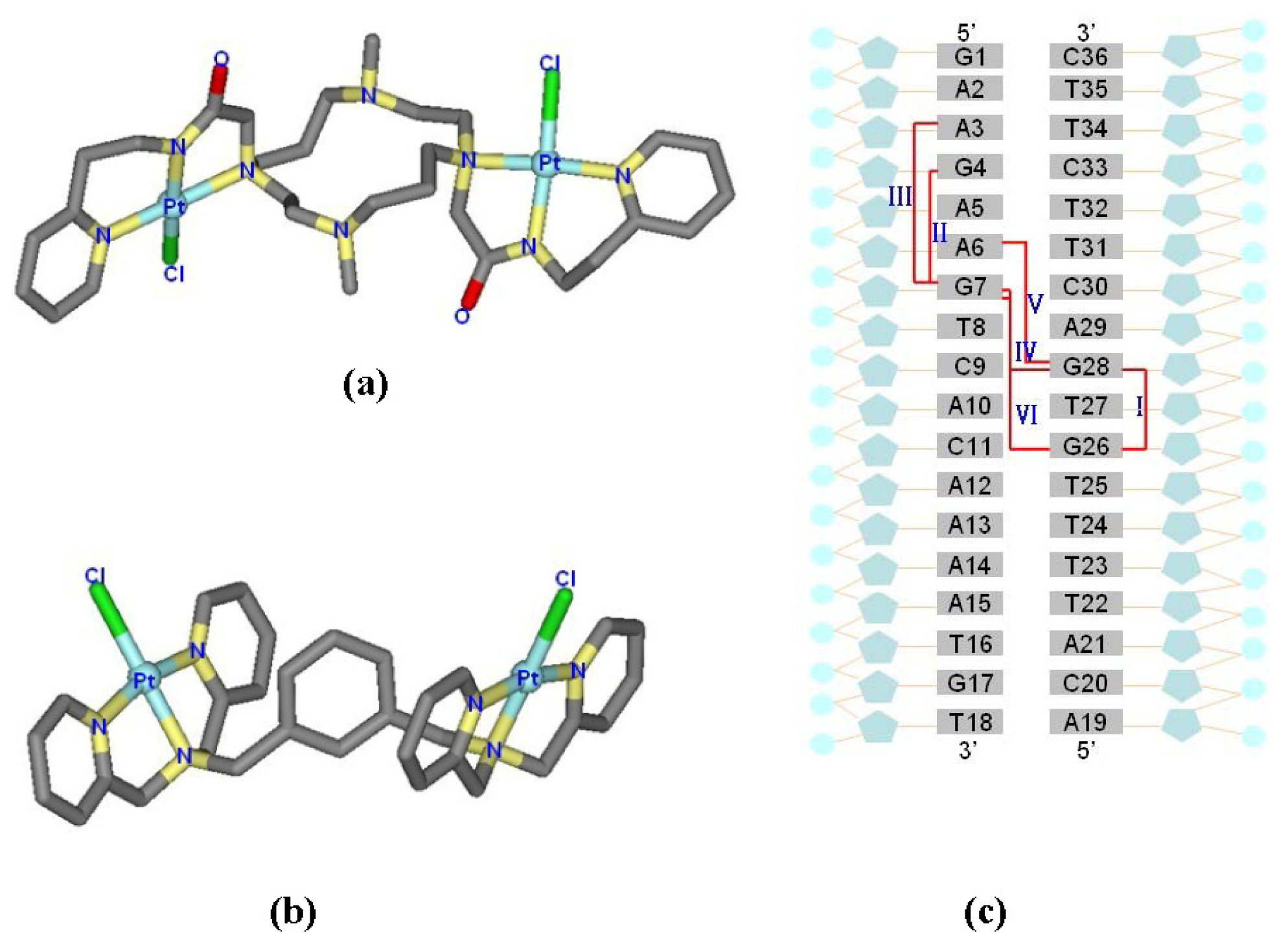
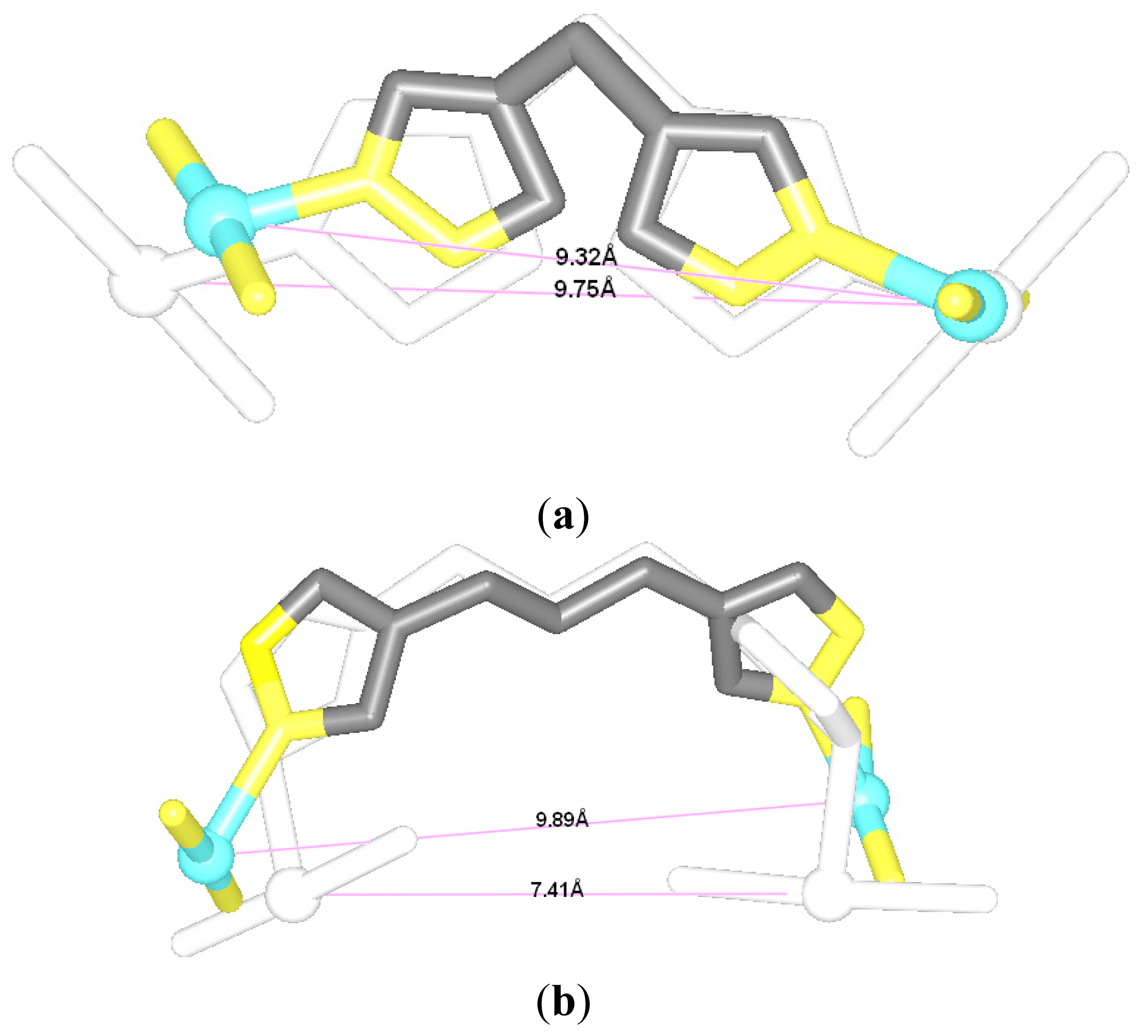
2.1. Conformational Characteristics and Flexibility of [Pt2(DTBPA)Cl2](II) and [Pt2(TPXA)Cl2](II)
2.2. Relationship between the Platinum–Platinum Distances in the Platinum(II) Agents and the N7–N7 Distances in the Pt2(DTBPA)-DNA and Pt2(TPXA)-DNA Adducts
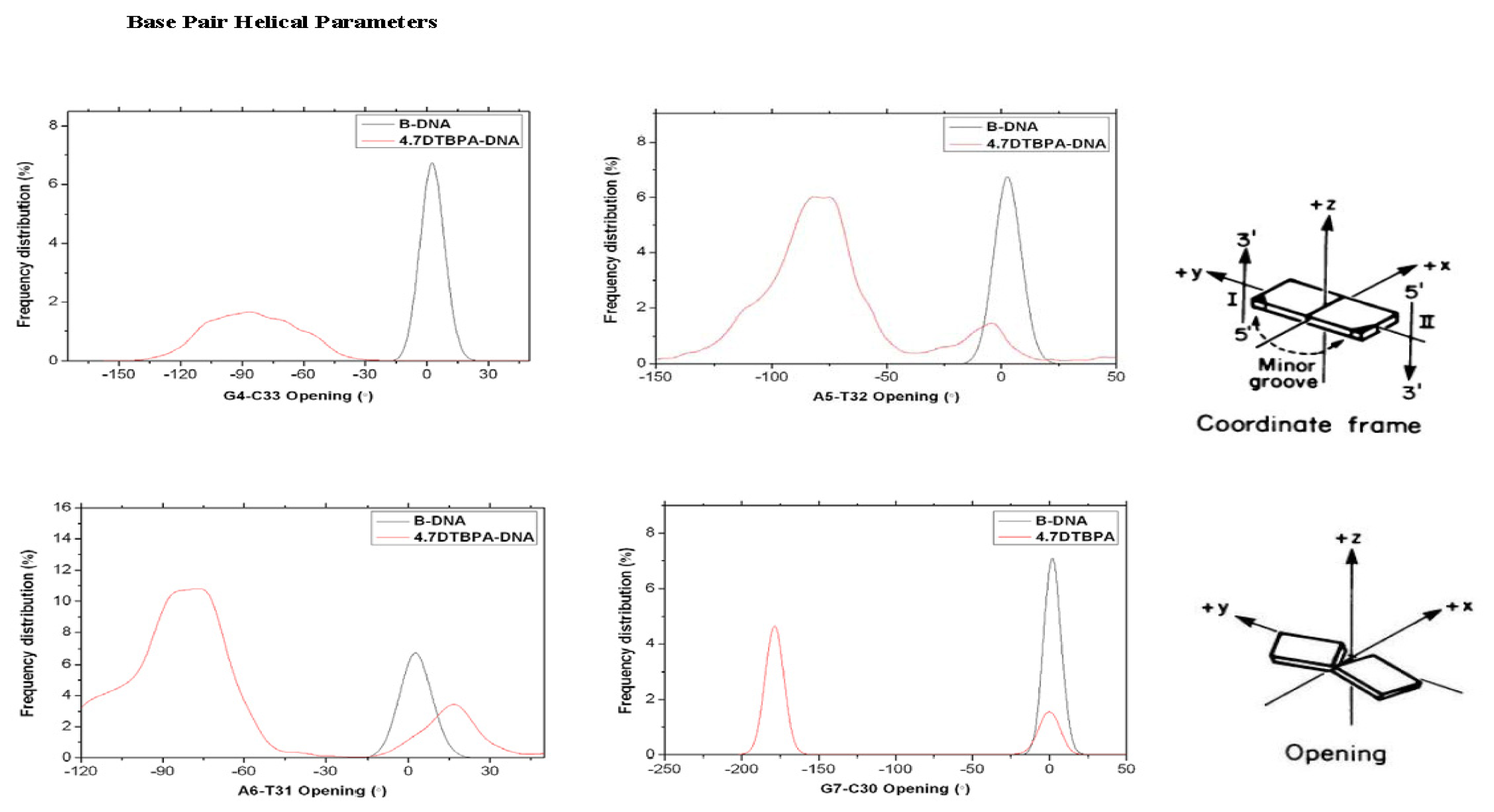
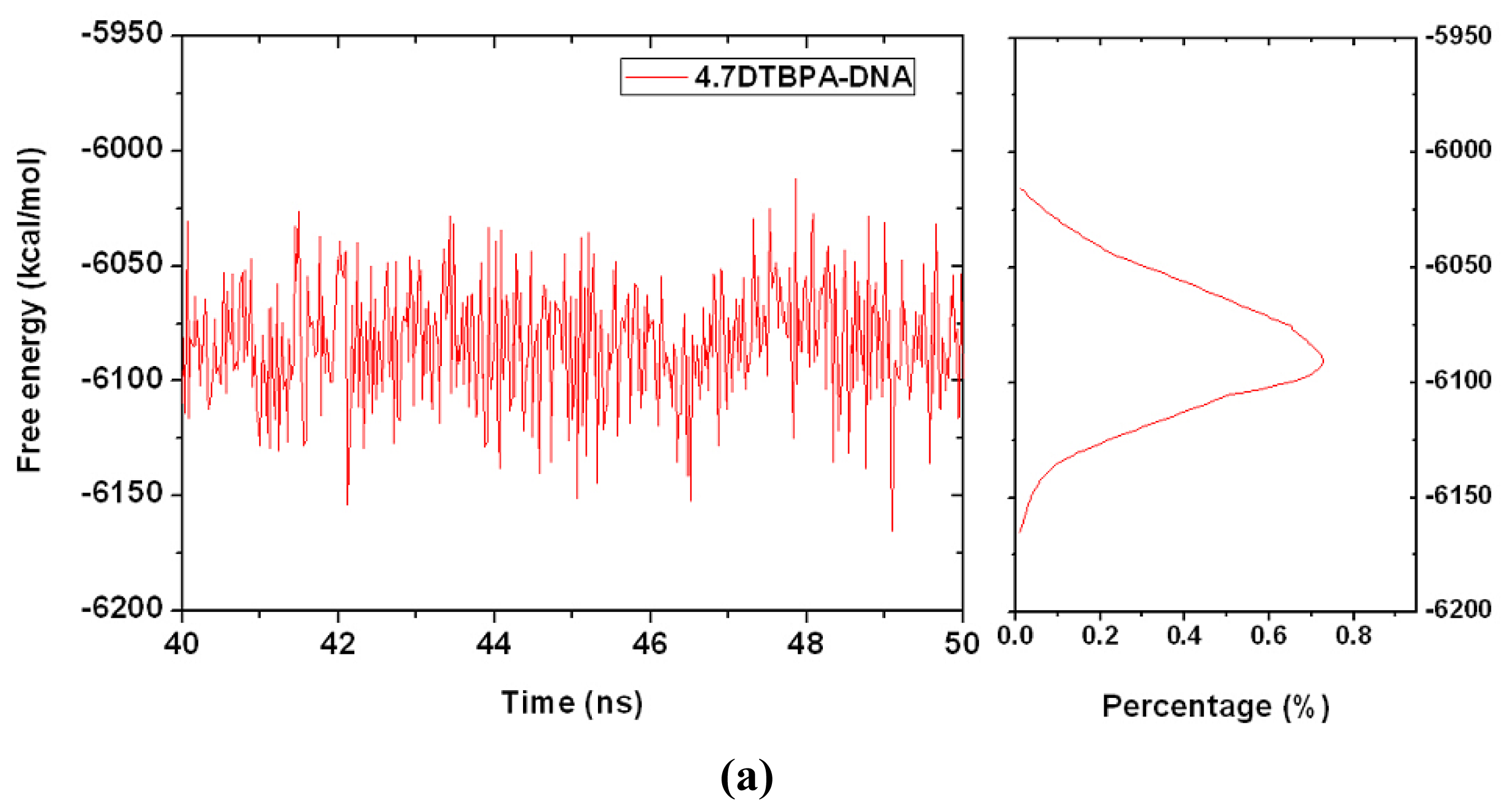
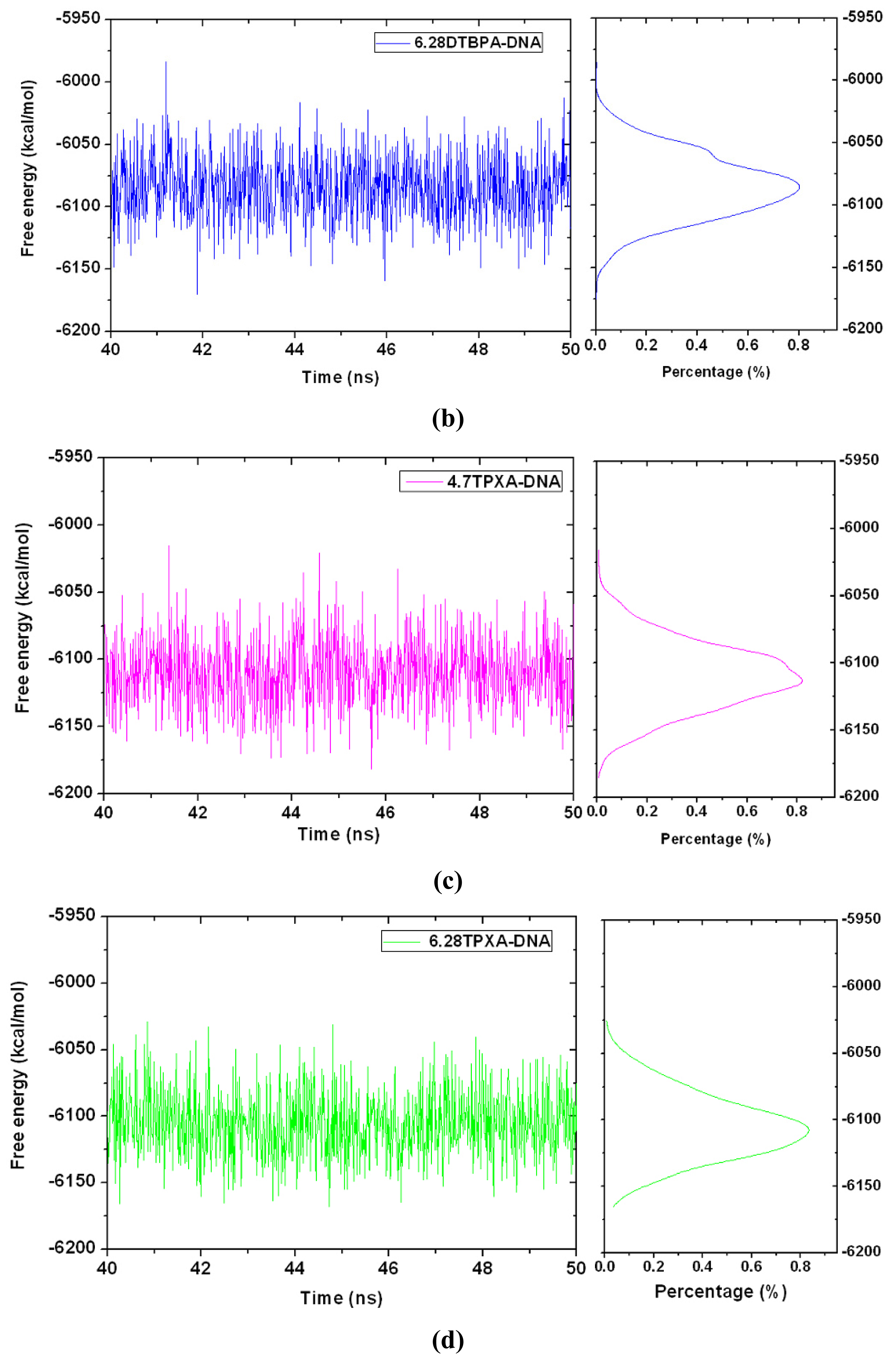
2.3. Relationship between the Flexibilities of the [Pt2(DTBPA)Cl2](II) and [Pt2(TPXA)Cl2](II) Agents and Their DNA Binding Interactions
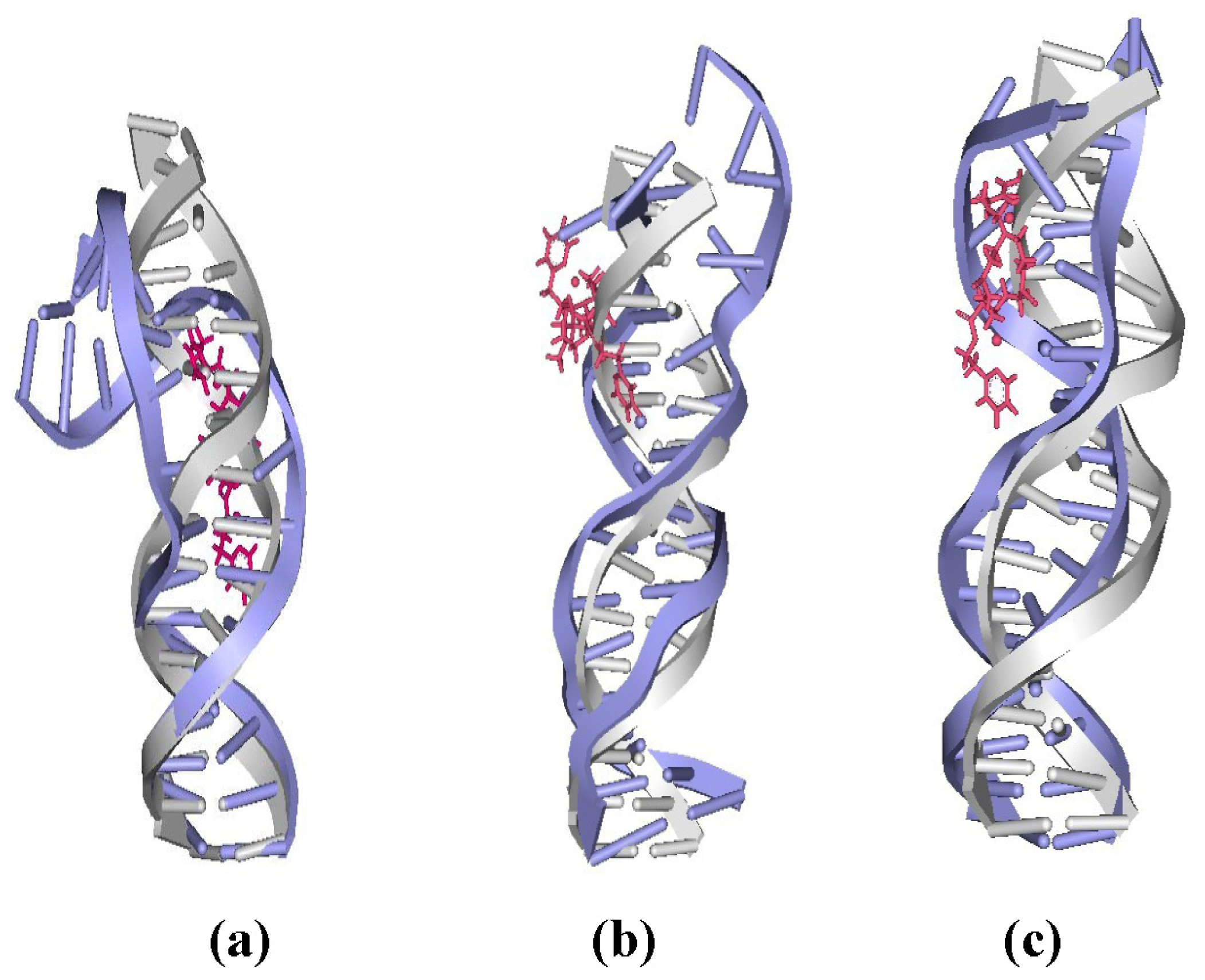
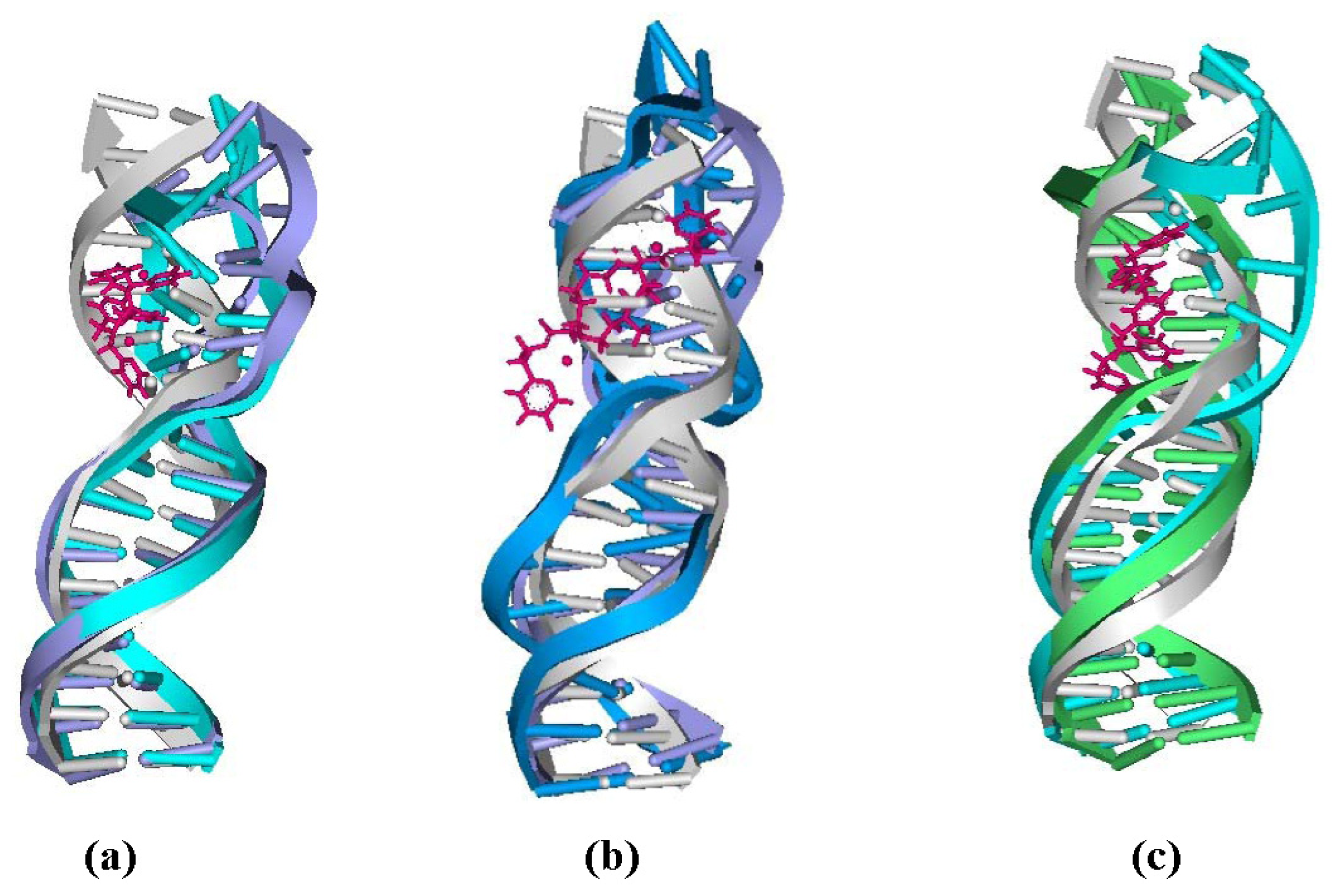
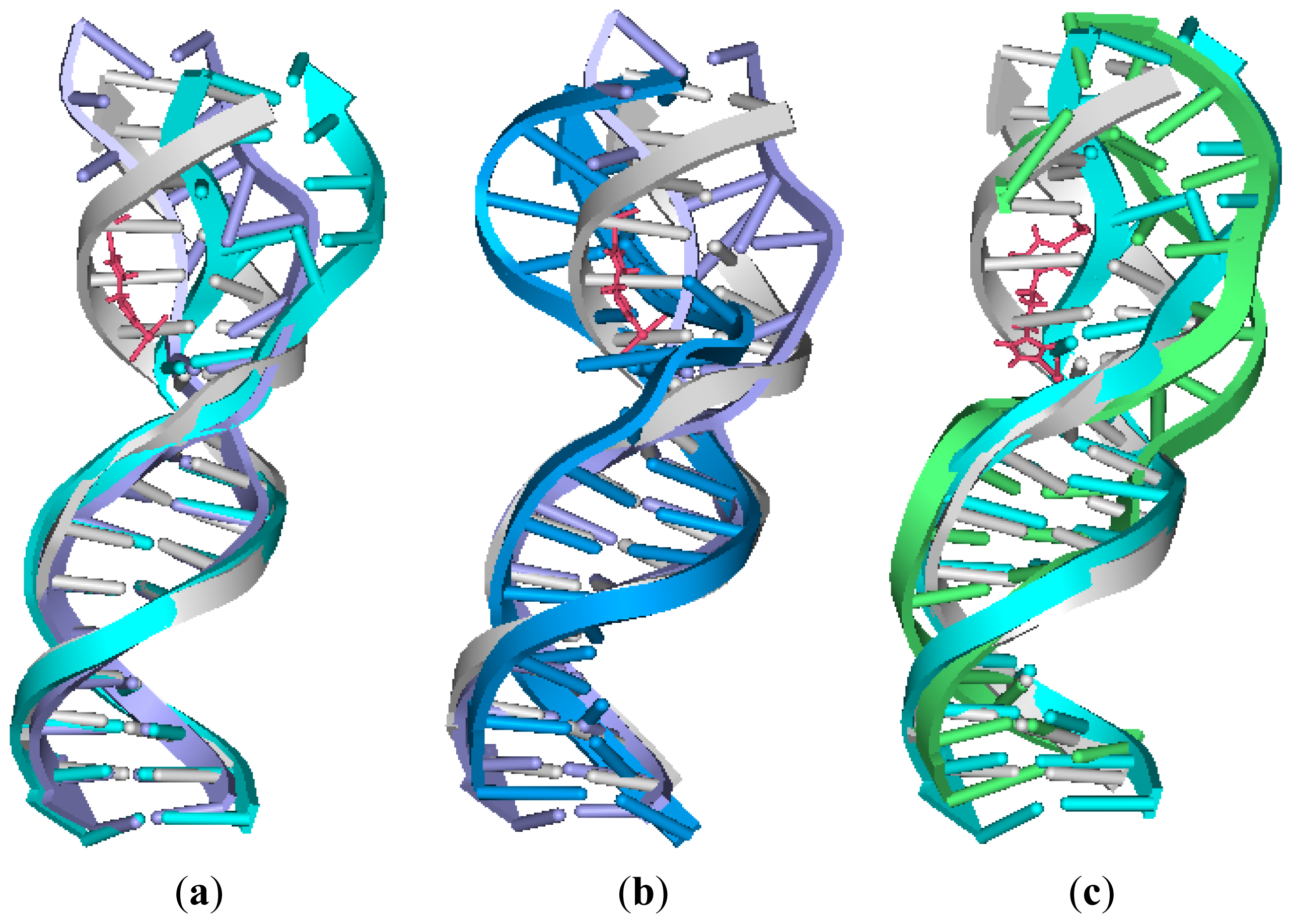
2.4. Relationship between the trans-[Pt2(DTBPA)Cl2](II)/cis-[Pt2(TPXA)Cl2](II) Configurations and the DNA Inter-/Intra-Strand Cross-Linking Modes
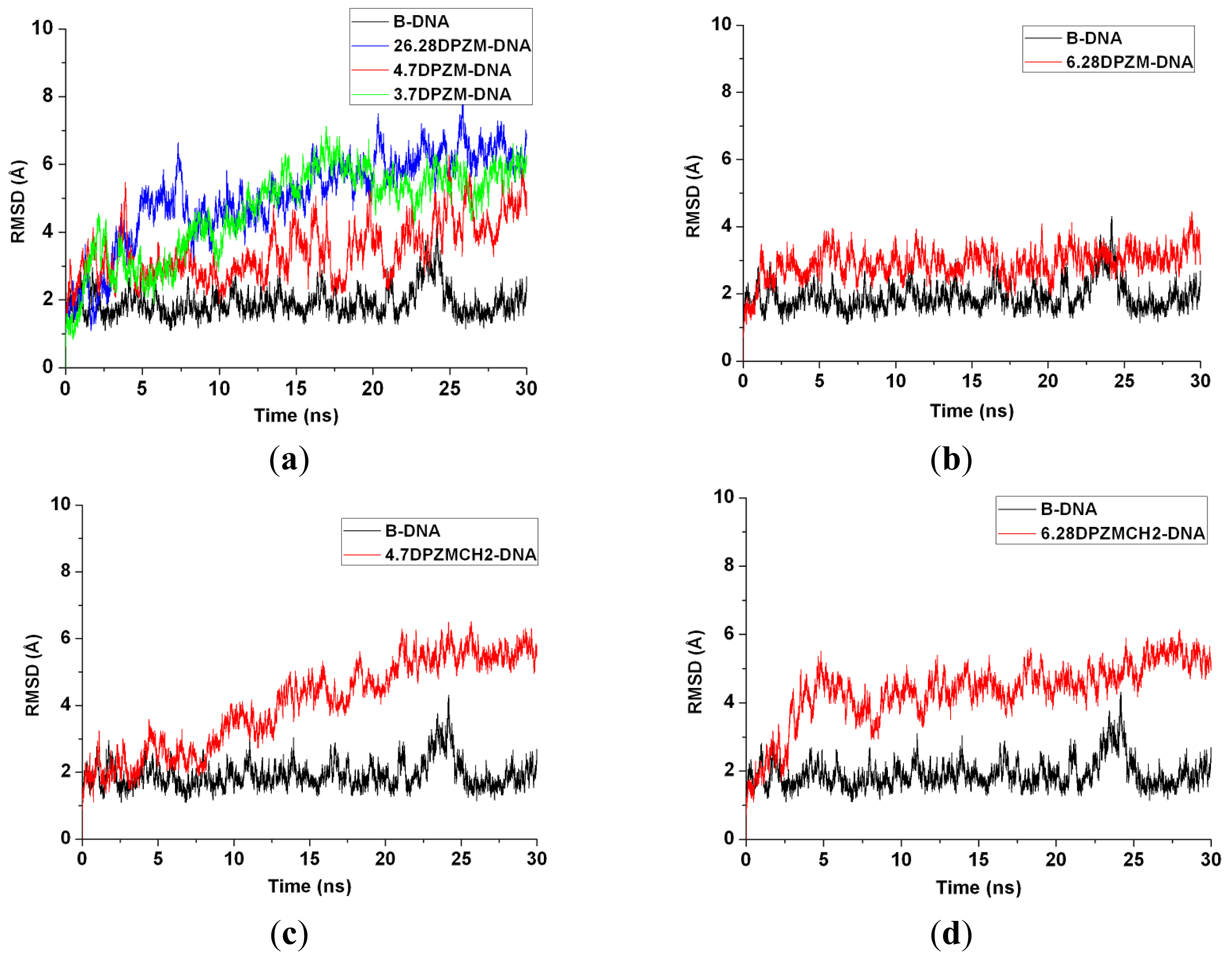
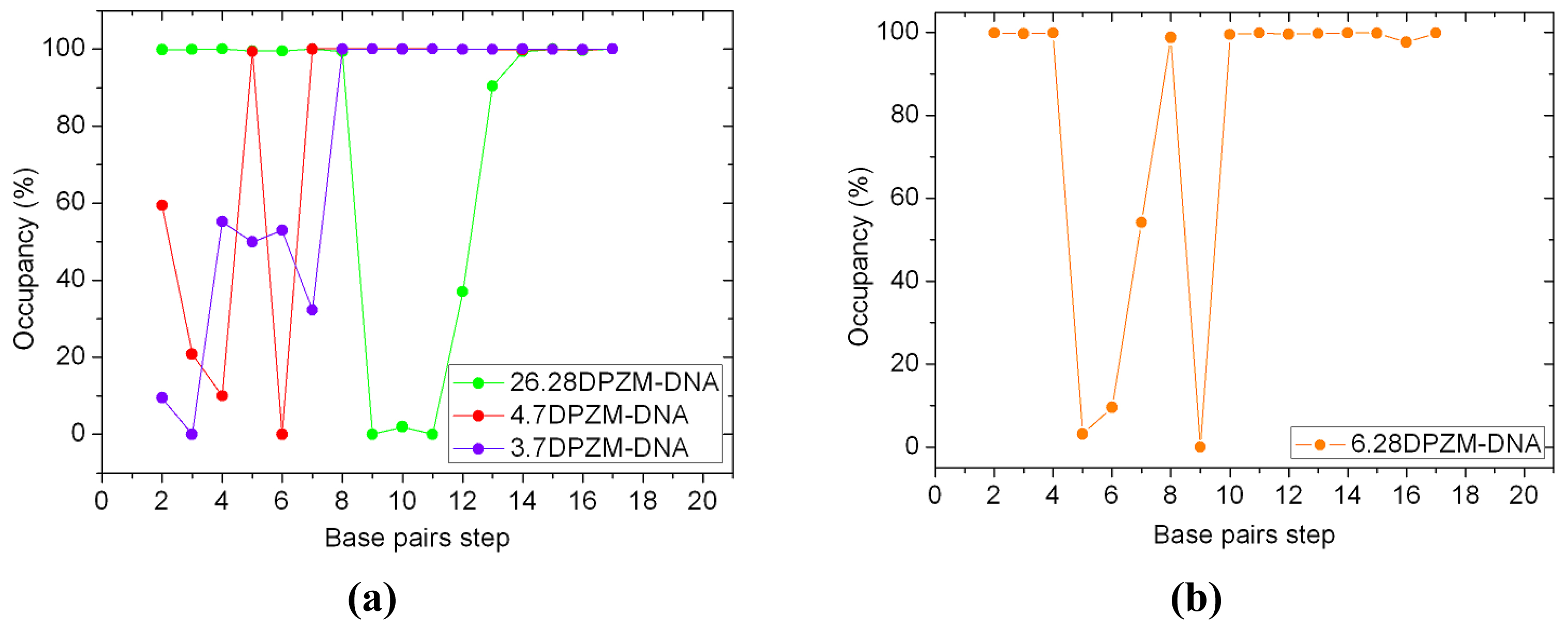
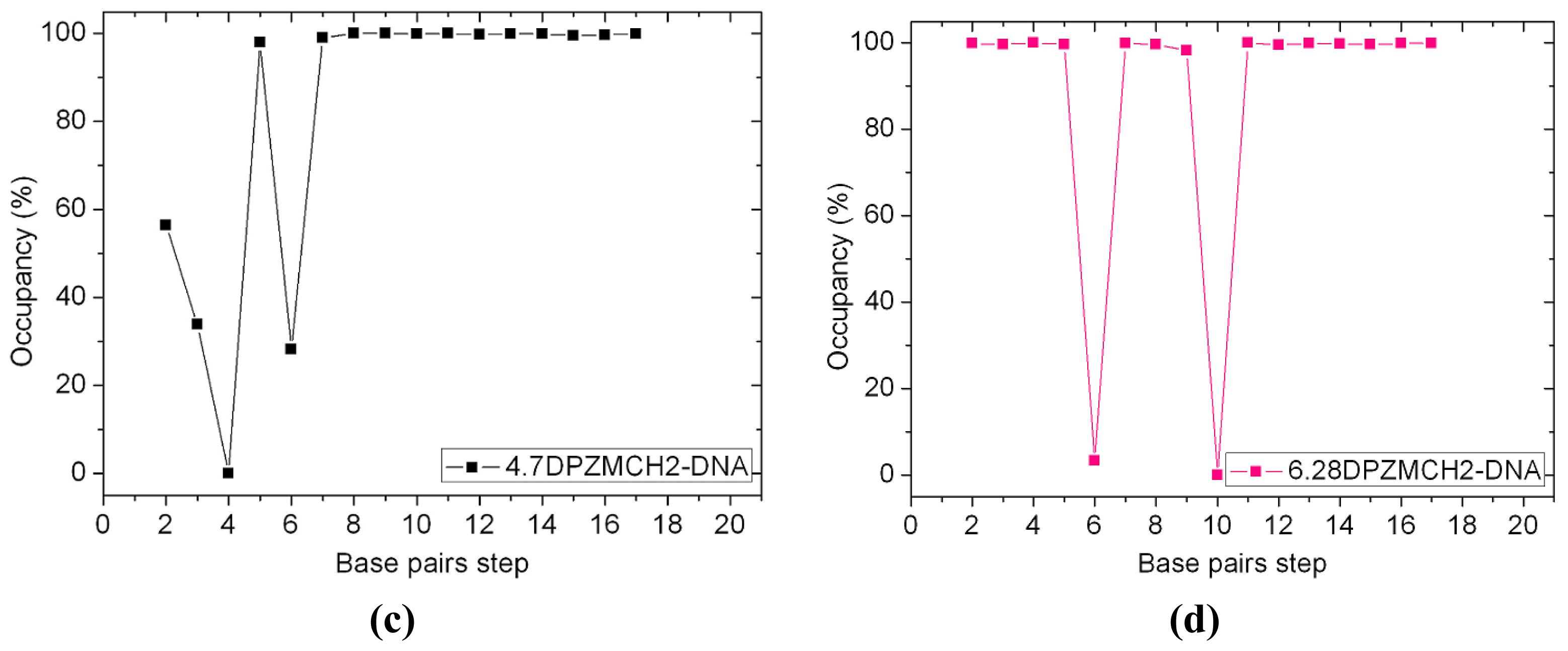
2.5. Configuration Characteristics of Two Additional Platinum Agents, trans-[Pt2(DPZM)Cl2](II) and cis-[Pt2(DPZMCH2)Cl2](II), Binding to DNA
3. Discussions
3.1. Principal Component Analysis of the Major Conformational Dynamics
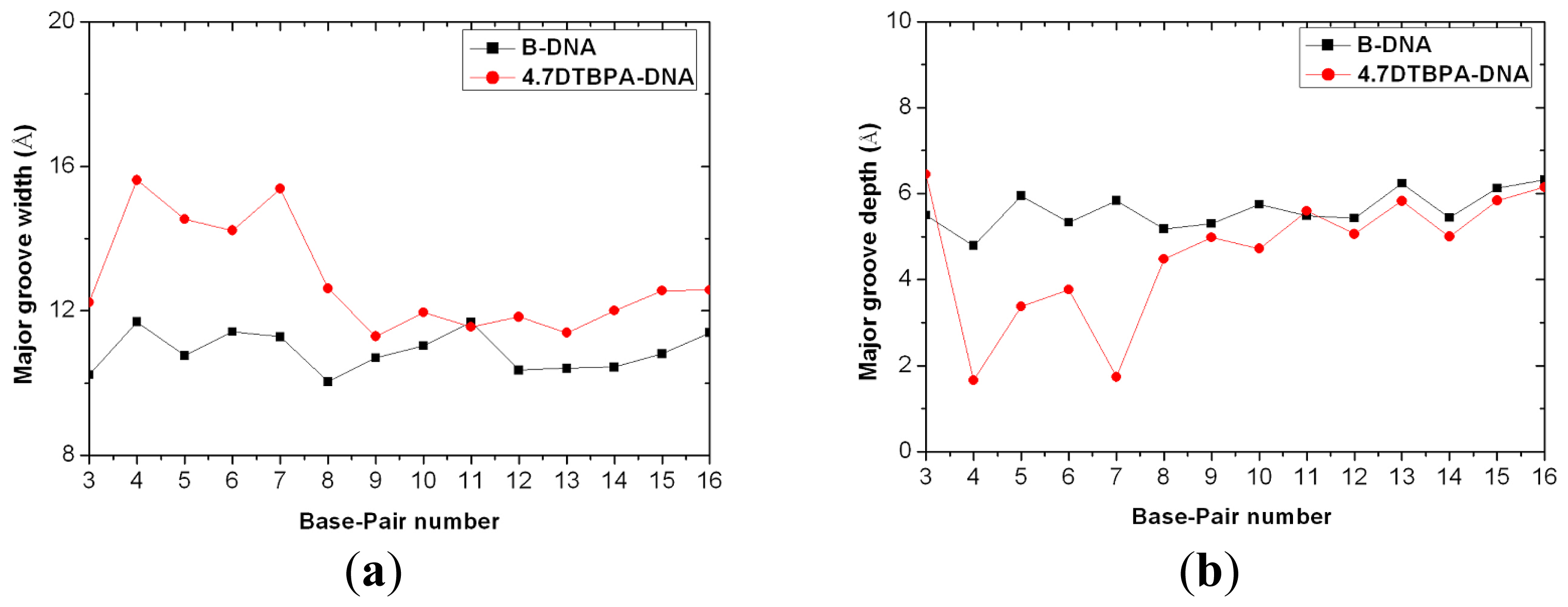
3.2. DNA Conformational Dynamics in the Platinum-DNA Adducts
4. Models and Methods
4.1. Starting Structures
4.2. Force Field Parameter Preparation
4.3. Molecular Dynamics Simulations and Trajectory Analysis
4.4. Principal Component Analysis
4.5. DNA Groove Parameter Analyses of Trajectories
5. Conclusions
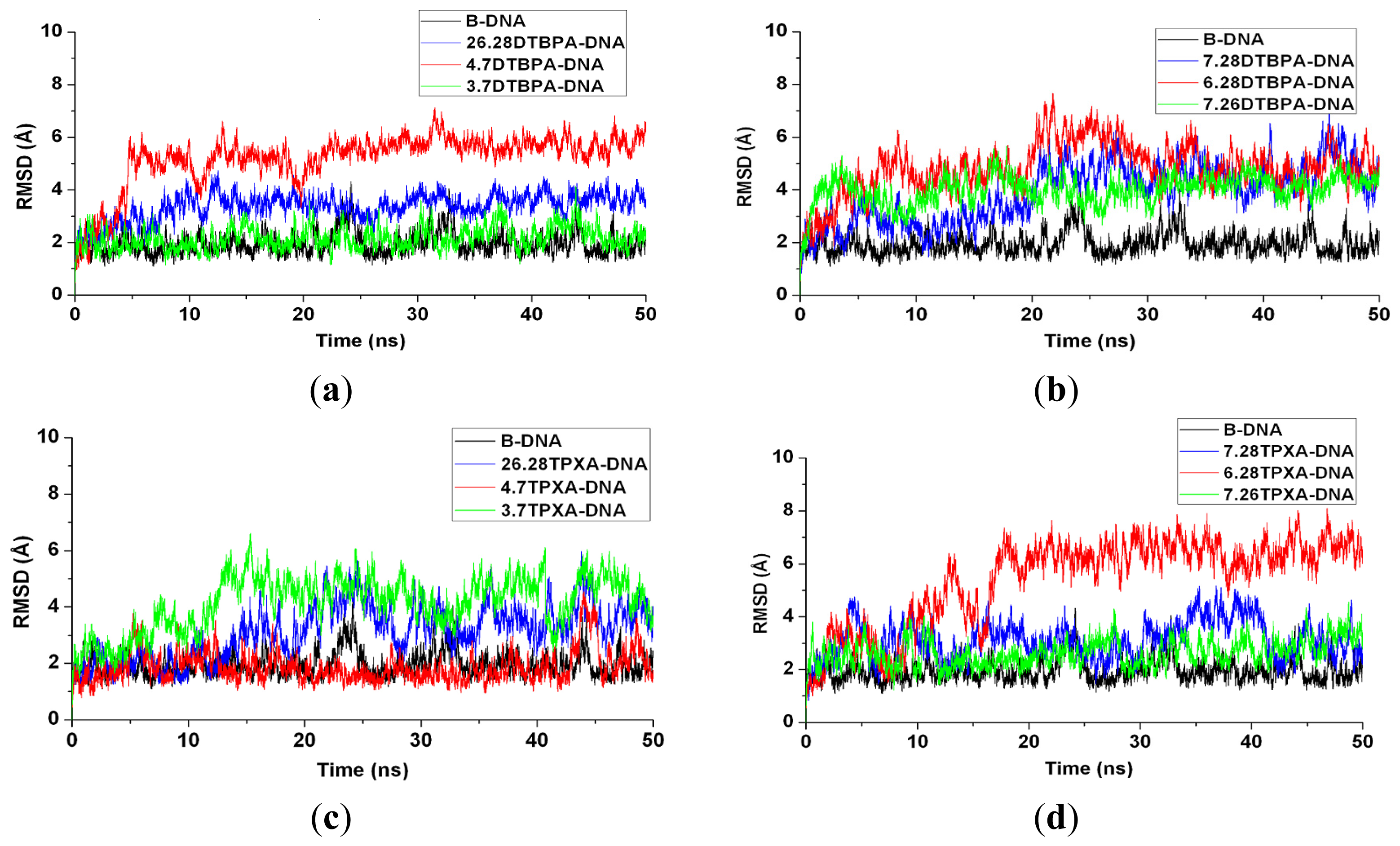
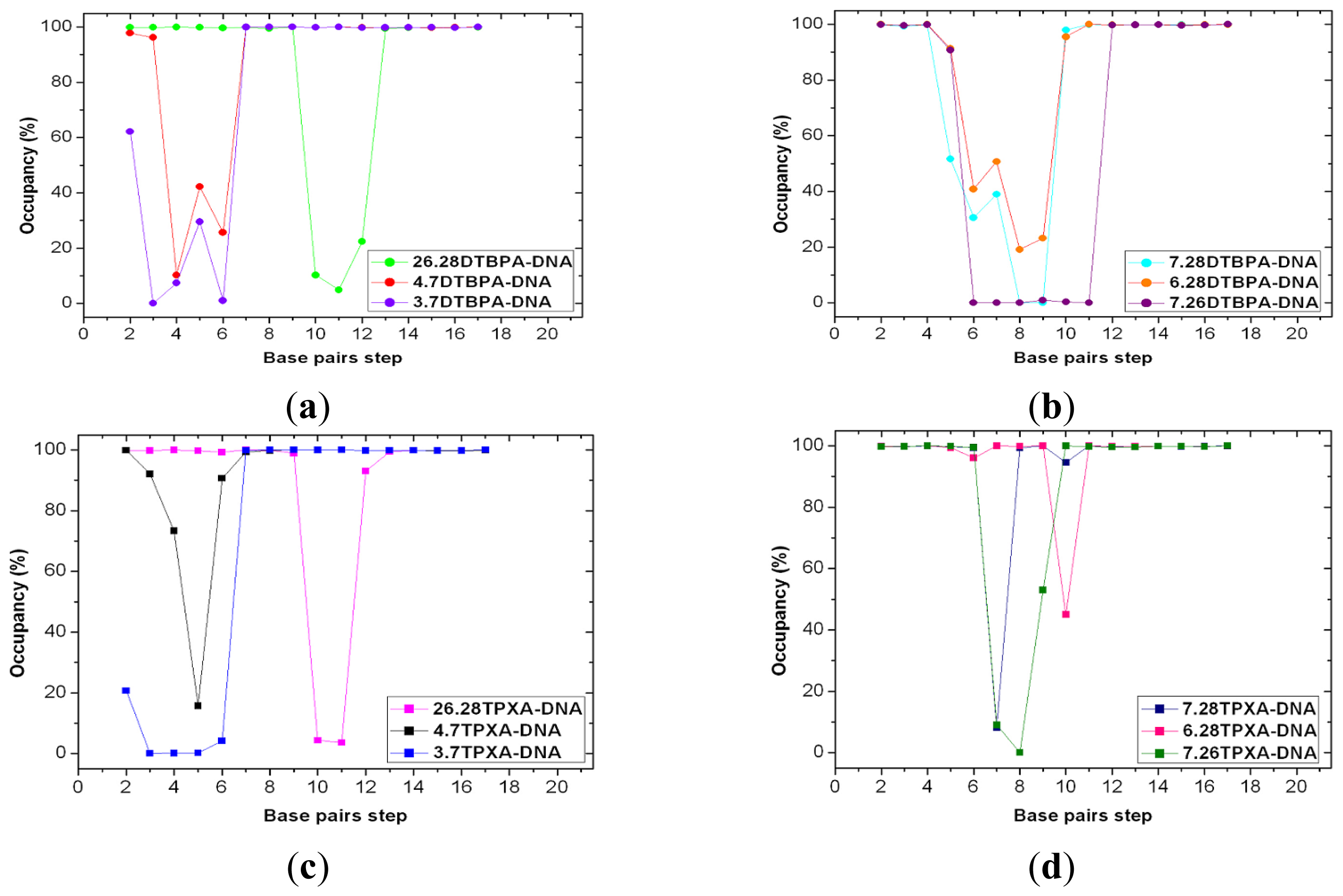
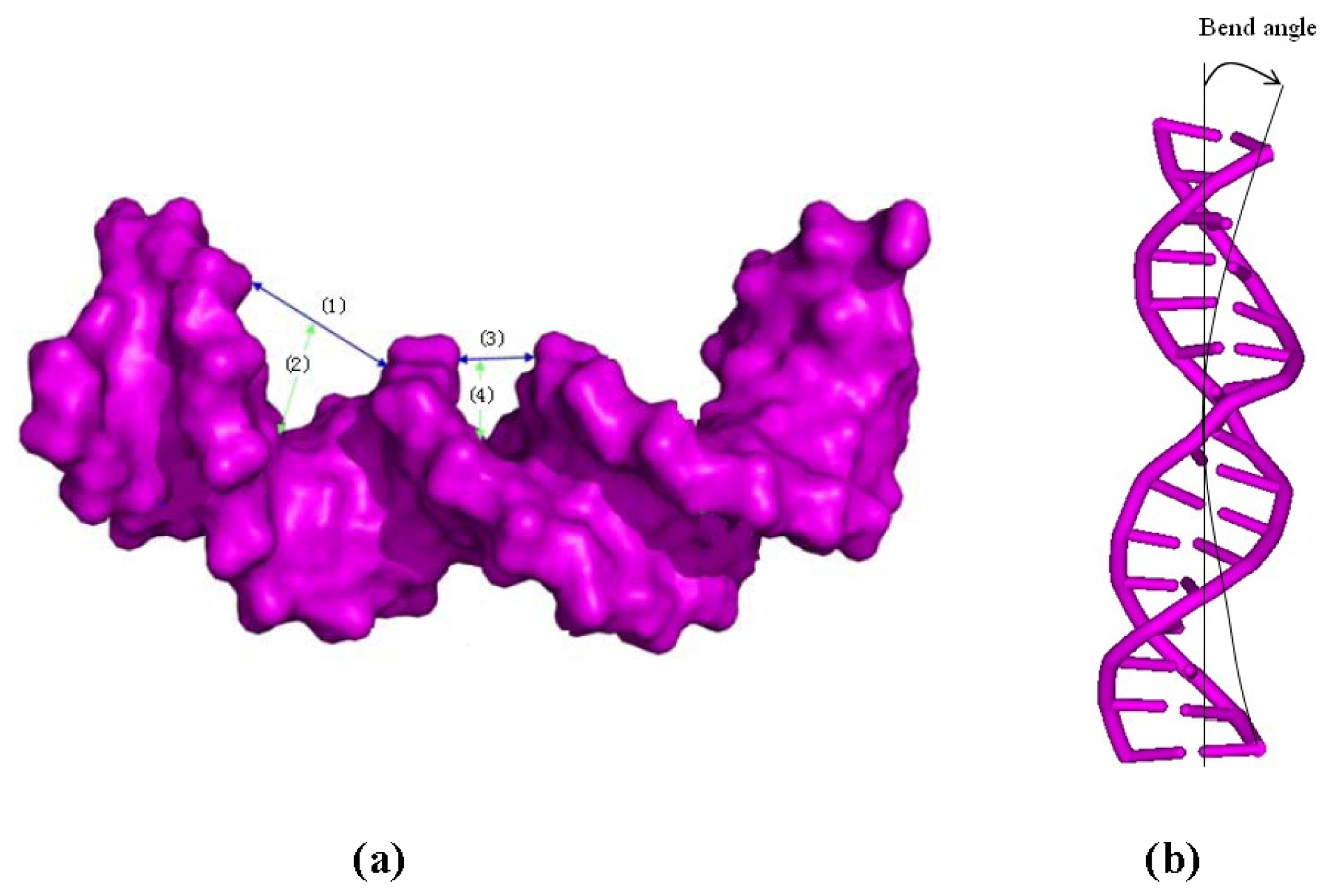
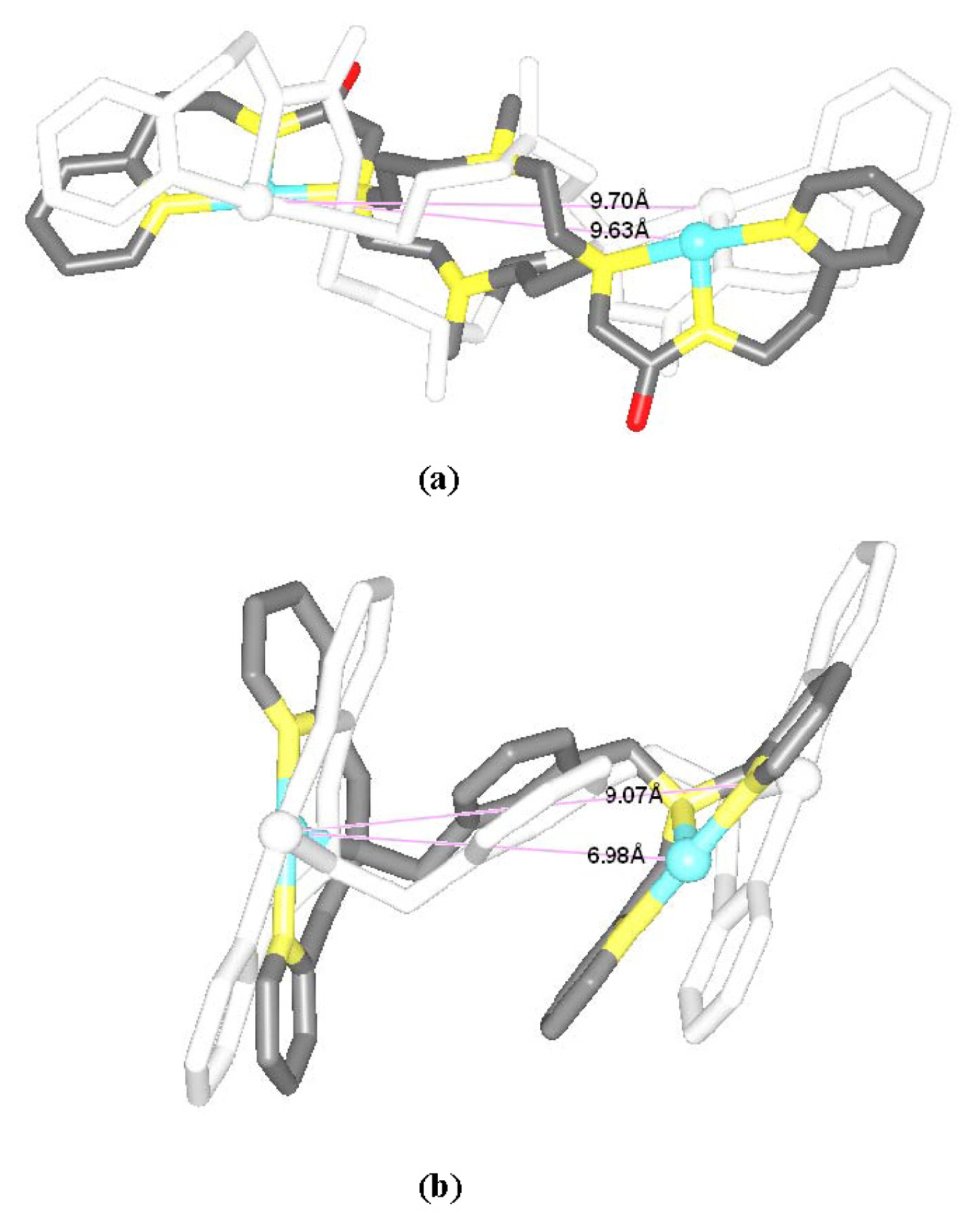


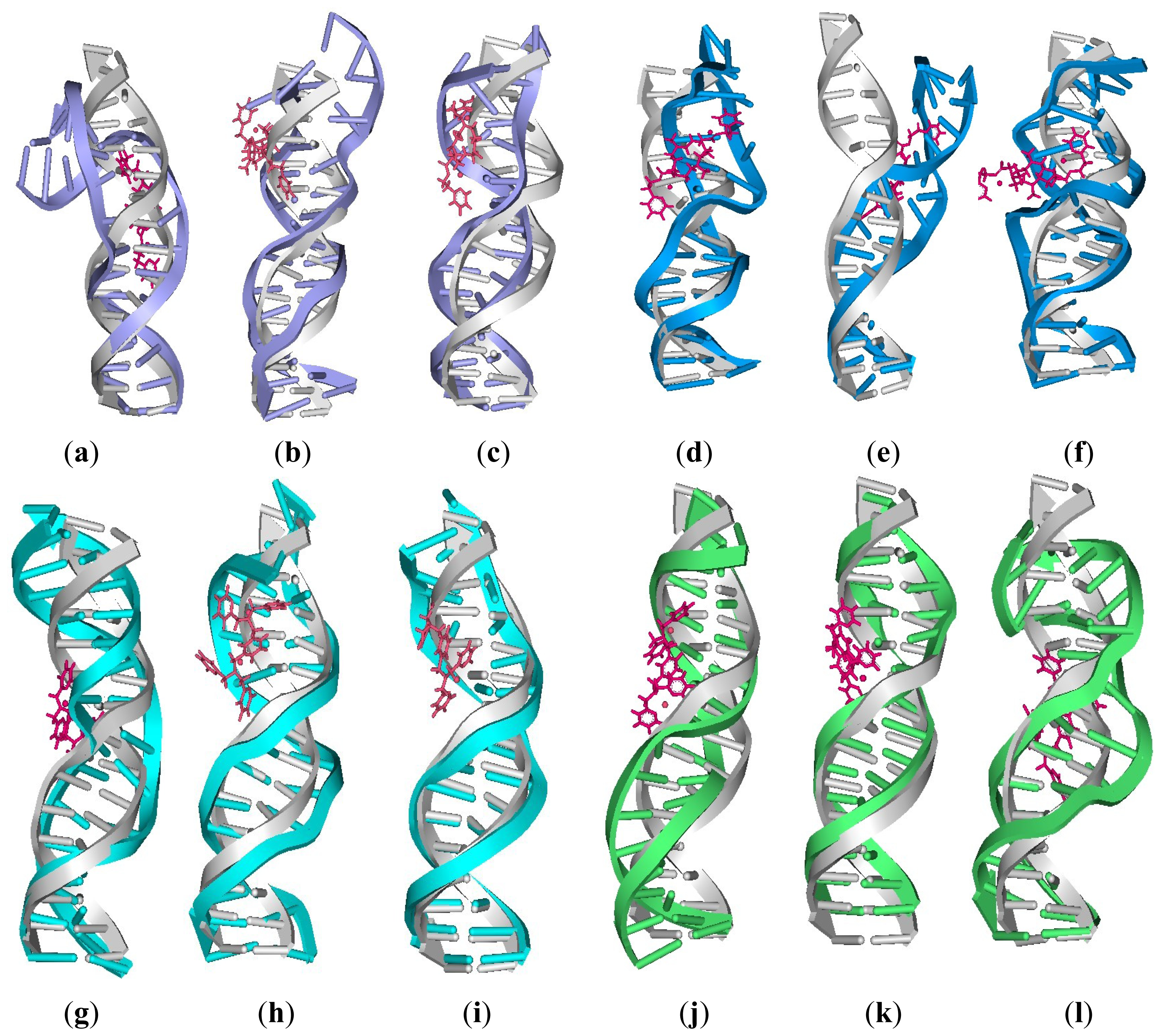
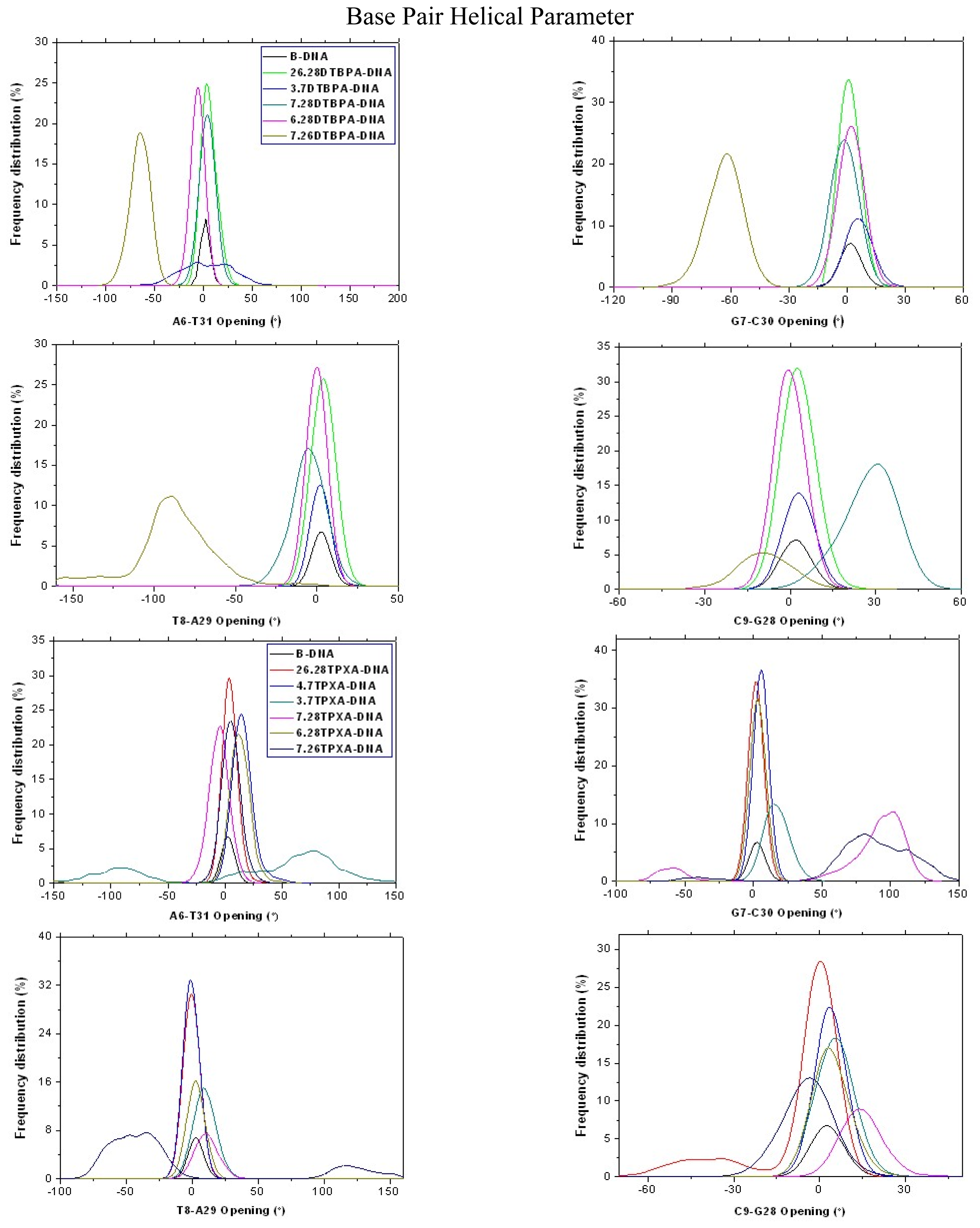
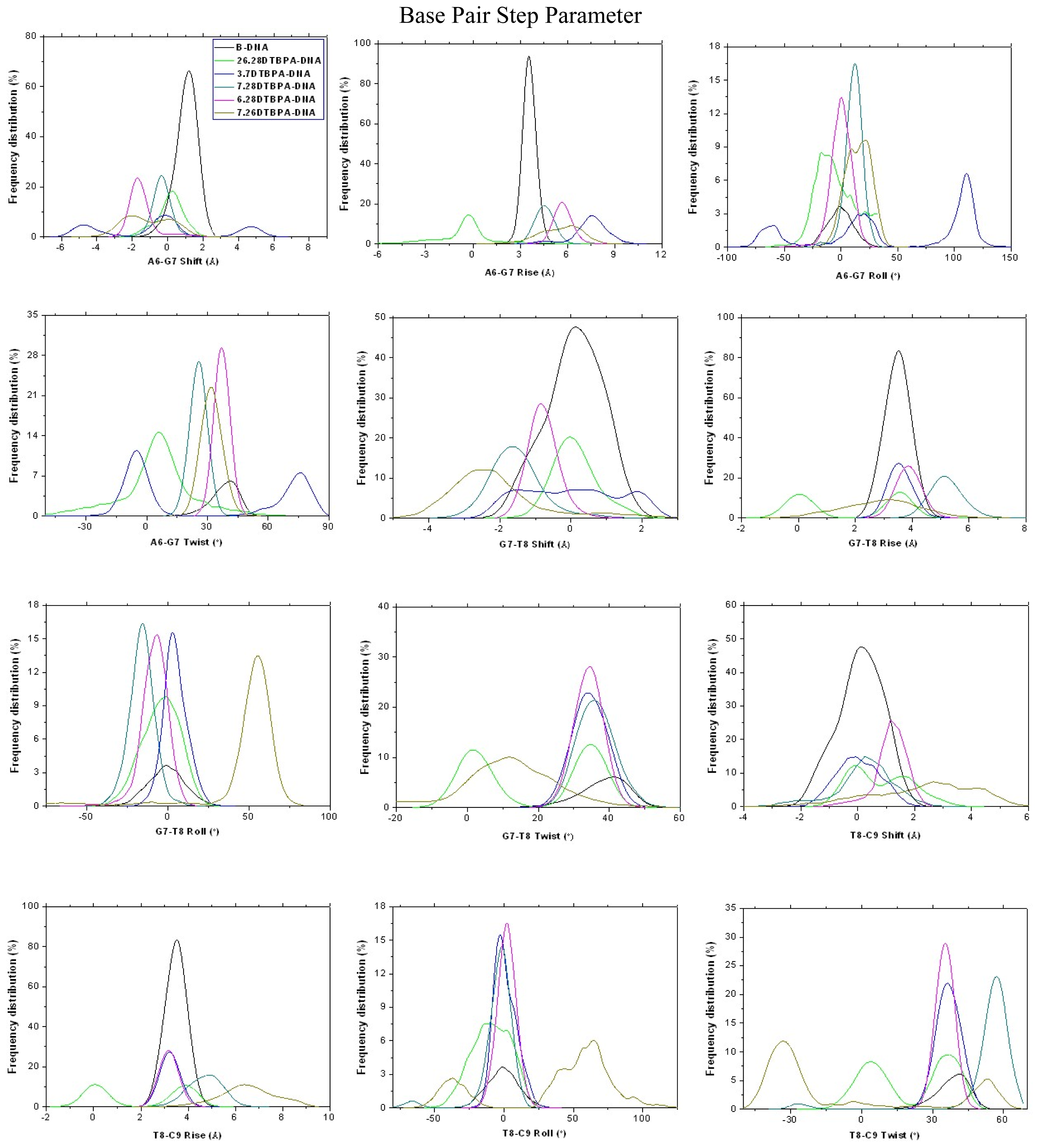

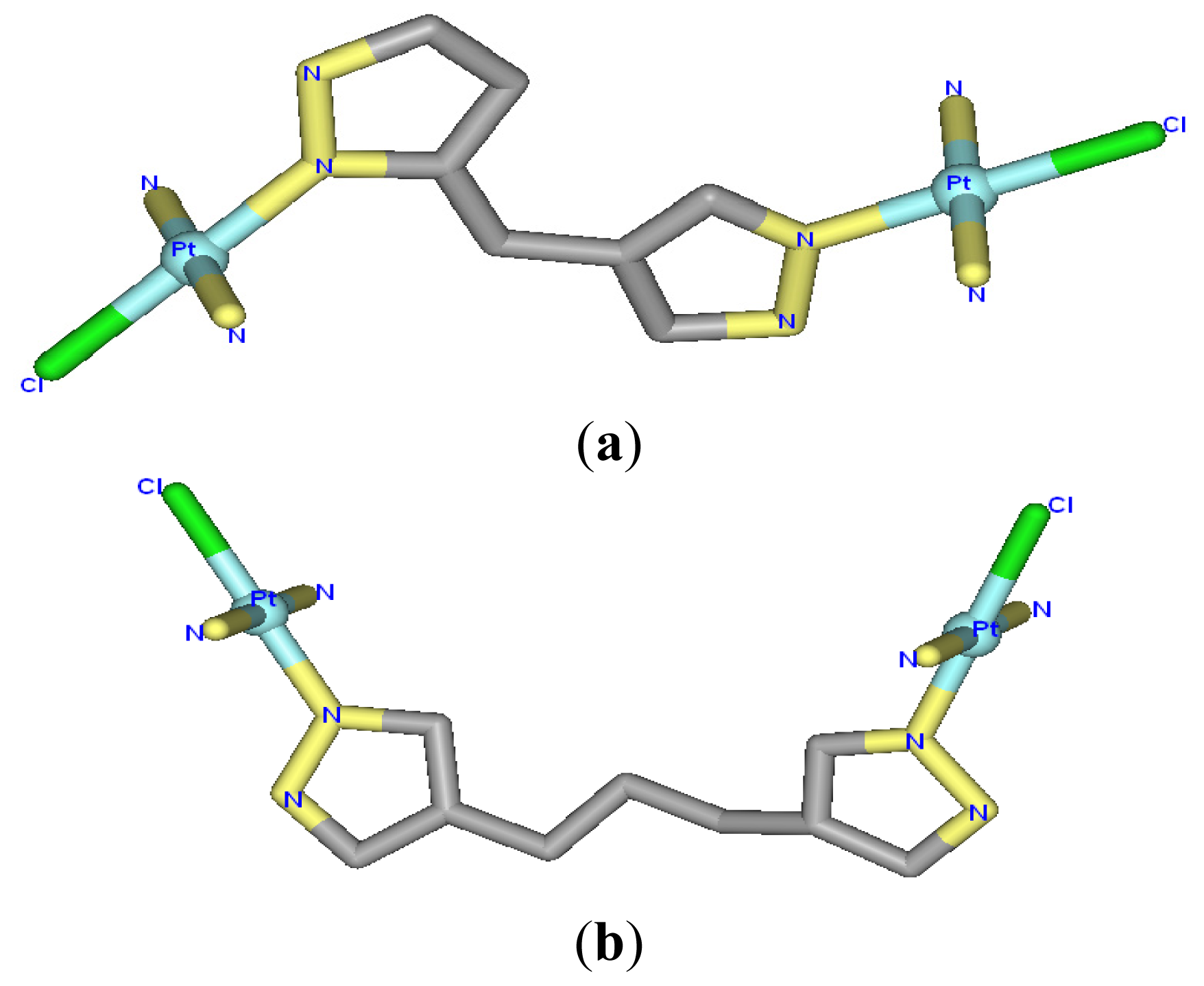
| Model | Major groove width (Å) | Major groove depth (Å) | Minor groove width (Å) | Minor groove depth (Å) | Bend (°) |
|---|---|---|---|---|---|
| 26.28DTBPA-DNA | 12.47 | 3.28 | 5.65 | 4.93 | 35.48 |
| 4.7DTBPA-DNA | 13.72 | 3.22 | 5.95 | 5.03 | 30.70 |
| 3.7DTBPA-DNA | 13.09 | 4.94 | 6.76 | 4.15 | 48.62 |
| 7.28DTBPA-DNA | 14.85 | 2.59 | 6.21 | 5.08 | 53.81 |
| 6.28DTBPA-DNA | 15.76 | 2.46 | 6.12 | 4.29 | 49.91 |
| 7.26DTBPA-DNA | 13.66 | 4.62 | 5.03 | 4.85 | 61.94 |
| 26.28TPXA-DNA | 12.27 | 5.70 | 5.91 | 5.04 | 27.20 |
| 4.7TPXA-DNA | 12.43 | 7.01 | 6.81 | 3.71 | 26.18 |
| 3.7TPXA-DNA | 10.52 | 6.18 | 6.53 | 3.27 | 49.15 |
| 7.28TPXA-DNA | 12.95 | 6.89 | 5.52 | 3.83 | 30.58 |
| 6.28TPXA-DNA | 11.98 | 5.73 | 6.04 | 4.28 | 27.15 |
| 7.26TPXA-DNA | 9.73 | 6.40 | 6.35 | 3.97 | 50.57 |
| B-DNA | 11.81 | 5.84 | 5.87 | 4.64 | 15.02 |
| Model | Pt–Pt distance (Å) | N7–N7 distance | Δ(Pt–N7) (Å) | Δ(N7–N7) (Å) | |
|---|---|---|---|---|---|
| B-DNA (Å) | Platinum-DNA adduct (Å) | ||||
| 3.7DTBPA-DNA | 9.70 | 14.81 | 11.05 | 5.11 | 3.76 |
| 4.7DTBPA-DNA | 9.70 | 11.60 | 11.22 | 1.90 | 0.38 |
| 26.28DTBPA-DNA | 9.70 | 7.63 | 9.85 | 2.07 | 2.22 |
| Model | PC1 | PC2 | PC3 | PCsum |
|---|---|---|---|---|
| 26.28DTBPA-DNA | 77.60 | 7.74 | 7.07 | 92.41 |
| 4.7DTBPA-DNA | 66.56 | 22.98 | 1.98 | 91.52 |
| 3.7DTBPA-DNA | 48.93 | 32.79 | 12.50 | 94.22 |
| 7.28DTBPA-DNA | 67.36 | 20.31 | 5.89 | 93.56 |
| 6.28DTBPA-DNA | 67.37 | 22.45 | 3.12 | 92.94 |
| 7.26DTBPA-DNA | 54.53 | 21.73 | 16.58 | 92.84 |
| 26.28TPXA-DNA | 60.40 | 26.49 | 4.22 | 91.11 |
| 4.7TPXA-DNA | 58.00 | 24.51 | 7.66 | 90.17 |
| 3.7TPXA-DNA | 63.53 | 19.50 | 8.80 | 91.83 |
| 7.28TPXA-DNA | 62.46 | 19.07 | 8.45 | 89.98 |
| 6.28TPXA-DNA | 40.14 | 331.45 | 116.72 | 888.31 |
| 7.26TPXA-DNA | 50.27 | 24.79 | 15.20 | 90.26 |
| Model | Major groove width (Å) | Major groove depth (Å) | Minor groove width (Å) | Minor groove depth (Å) | Bend(°) |
|---|---|---|---|---|---|
| 26.28DPZM-DNA | 13.27 | 6.51 | 4.67 | 5.64 | 43.91 |
| 4.7DPZM-DNA | 12.95 | 4.49 | 6.42 | 4.14 | 30.88 |
| 3.7DPZM-DNA | 13.11 | 4.35 | 6.23 | 4.36 | 34.94 |
| 6.28DPZM-DNA | 15.40 | 2.51 | 4.16 | 3.79 | 48.73 |
| 4.7DPZMCH2-DNA | 12.41 | 6.62 | 6.04 | 3.98 | 21.92 |
| 6.28DPZMCH2-DNA | 11.47 | 6.23 | 6.51 | 4.16 | 19.24 |
| B-DNA | 11.81 | 5.84 | 5.87 | 4.64 | 15.02 |
| Model | Pt–Pt distance (Å) | N7–N7 distance | Δ(Pt–N7) (Å) | Δ(N7–N7) (Å) | |
|---|---|---|---|---|---|
| B-DNA (Å) | Platinum-DNA adduct (Å) | ||||
| 3.7DPZM-DNA | 9.75 | 14.81 | 12.80 | 5.06 | 2.01 |
| 4.7DPZM-DNA | 9.75 | 11.60 | 11.47 | 1.85 | 0.13 |
| 26.28DPZM-DNA | 9.75 | 7.63 | 11.07 | 2.12 | 3.44 |
Acknowledgments
Appendix
Molecular Dynamics Simulation Protocols
Principal Component Analysis
DNA Groove Parameter Analyses of the Trajectories
Conflicts of Interest
References
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov 2005, 4, 307–320. [Google Scholar]
- Jung, Y.; Lippard, S.J. Direct cellular responses to platinum-induced DNA damage. Chem. Rev 2007, 107, 1387–1407. [Google Scholar]
- Steer, C.B.; Chrystal, K.; Cheong, K.A.; Galani, E.; Marx, G.M.; Strickland, A.H.; Yip, D.; Lofts, F.; Gallagher, C.; Thomas, H.; et al. Gemcitabine and oxaliplatin followed by paclitaxel and carboplatin as first line therapy for patients with suboptimally debulked, advanced epithelial ovarian cancer. A phase II trial of sequential doublets. The GO-First Study. Gynecol. Oncol 2006, 103, 439–445. [Google Scholar]
- McWhinney, S.R.; Goldberg, R.M.; McLeod, H.L. Platinum neurotoxicity pharmacogenetics. Mol. Cancer Ther 2009, 8, 10–16. [Google Scholar]
- Kormos, B.L.; Benitex, Y.; Baranger, A.M.; Beveridge, D.L. Affinity and specificity of protein U1A-RNA complex formation based on an additive component free energy model. J. Mol. Biol 2007, 371, 1405–1419. [Google Scholar]
- Reedijk, J. New clues for platinum antitumor chemistry: Kinetically controlled metal binding to DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 3611–3616. [Google Scholar]
- Park, G.Y.; Wilson, J.J.; Song, Y.; Lippard, S.J. Phenanthriplatin, a monofunctional DNA-binding platinum anticancer drug candidate with unusual potency and cellular activity profile. Proc. Natl. Acad. Sci. USA 2012, 109, 11987–11992. [Google Scholar]
- Pizarro, A.M.; Sadler, P.J. Unusual DNA binding modes for metal anticancer complexes. Biochimie 2009, 91, 1198–1211. [Google Scholar]
- Silverman, A.P.; Bu, W.; Cohen, S.M.; Lippard, S.J. 2.4 Å Crystal structure of the asymmetric platinum complex {Pt (ammine)(cyclohexylamine)}2+ bound to a dodecamer DNA duplex. J. Biol. Chem 2002, 277, 49743–49749. [Google Scholar]
- Zhu, Y.; Wang, Y.; Chen, G. Differences in conformational dynamics of [Pt3 (HPTAB)]6+-DNA adducts with various cross-linking modes. Nucleic Acids Res 2009, 37, 5930–5942. [Google Scholar]
- Kasparkova, J.; Mackay, F.S.; Brabec, V.; Sadler, P.J. Formation of platinated GG cross-links on DNA by photoactivation of a platinum(IV) azide complex. J. Biol. Inorg. Chem 2003, 8, 741–745. [Google Scholar]
- Klein, A.V.; Hambley, T.W. Platinum drug distribution in cancer cells and tumors. Chem. Rev 2009, 109, 4911–4920. [Google Scholar]
- Agarwal, R.; Kaye, S.B. Ovarian cancer: Strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer 2003, 3, 502–516. [Google Scholar]
- Ramachandran, S.; Temple, B.R.; Chaney, S.G.; Dokholyan, N.V. Structural basis for the sequence-dependent effects of platinum-DNA adducts. Nucleic Acids Res 2009, 37, 2434–2448. [Google Scholar]
- Shah, N.; Dizon, D.S. New-generation platinum agents for solid tumors. Future Oncol 2009, 5, 33–42. [Google Scholar]
- Kelland, L. Broadening the clinical use of platinum drug-based chemotherapy with new analogues. Expert Opin. Invest. Drugs 2007, 16, 1009–1021. [Google Scholar]
- Hegmans, A.; Berners-Price, S.J.; Davies, M.S.; Thomas, D.S.; Humphreys, A.S.; Farrell, N. Long range 1,4 and 1,6-interstrand cross-links formed by a trinuclear platinum complex. Minor groove preassociation affects kinetics and mechanism of cross-link formation as well as adduct structure. J. Am. Chem. Soc 2004, 126, 2166–2180. [Google Scholar]
- Mambanda, A.; Jaganyi, D.; Hochreuther, S.; van Eldik, R. Tuning the reactivity of chelated dinuclear Pt(II) complexes through a flexible diamine linker. A detailed kinetic and mechanistic study. Dalton Trans 2010, 39, 3595–3608. [Google Scholar]
- Zerzankova, L.; Kostrhunova, H.; Vojtiskova, M.; Novakova, O.; Suchankova, T.; Lin, M.X.; Guo, Z.J.; Kasparkova, J.; Brabec, V. Mechanistic insights into antitumor effects of new dinuclear cis Pt(II) complexes containing aromatic linkers. Biochem. Pharmacol 2010, 80, 344–351. [Google Scholar]
- Ruhayel, R.A.; Zgani, I.; Berners-Price, S.J.; Farrell, N.P. Solution studies of dinuclear polyamine-linked platinum-based antitumour complexes. Dalton Trans 2011, 40, 4147–4154. [Google Scholar]
- Kida, N.; Katsuda, Y.; Yoshikawa, Y.; Komeda, S.; Sato, T.; Saito, Y.; Chikuma, M.; Suzuki, M.; Imanaka, T.; Yoshikawa, K. Characteristic effect of an anticancer dinuclear platinum(II) complex on the higher-order structure of DNA. J. Biol. Inorg. Chem 2010, 15, 701–707. [Google Scholar]
- Zhu, J.H.; Lin, M.X.; Fan, D.M.; Wu, Z.Y.; Chen, Y.C.; Zhang, J.F.; Lu, Y.; Guo, Z.J. The role of bridging ligands in determining DNA-binding ability and cross-linking patterns of dinuclear platinum(II) antitumour complexes. Dalton Trans 2009, 38, 10889–10895. [Google Scholar]
- Farrell, N. Polynuclear platinum drugs. Met. Ions Biol. Syst 2004, 42, 251–296. [Google Scholar]
- Xu, Z.; Zhang, Y.; Xue, Z.; Yang, X.; Wu, Z.; Guo, Z. DNA-binding property and antitumor activity of a cyclam bridged dinuclear platinum (II) complex. Inorg. Chim. Acta 2009, 362, 2347–2352. [Google Scholar]
- Zhu, J.; Zhao, Y.; Zhu, Y.; Wu, Z.; Lin, M.; He, W.; Wang, Y.; Chen, G.; Dong, L.; Zhang, J.; et al. DNA cross-linking patterns induced by an antitumor-active trinuclear platinum complex and comparison with its dinuclear analogue. Chem.Eur. J 2009, 15, 5245–5253. [Google Scholar]
- Malina, J.; Kasparkova, J.; Farrell, N.P.; Brabec, V. Walking of antitumor bifunctional trinuclear PtII complex on double-helical DNA. Nucleic Acids Res 2011, 39, 720–728. [Google Scholar]
- Sharma, S.; Gong, P.; Temple, B.; Bhattacharyya, D.; Dokholyan, N.V.; Chaney, S.G. Molecular dynamic simulations of cisplatin-and oxaliplatin-d (GG) intrastand cross-links reveal differences in their conformational dynamics. J. Mol. Biol 2007, 373, 1123–1140. [Google Scholar]
- Arriagada, R.; Bergman, B.; Dunant, A.; le Chevalier, T.; Pignon, J.P.; Vansteenkiste, J. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N. Engl. J. Med 2004, 350, 351–360. [Google Scholar]
- Harris, A.L.; Yang, X.; Hegmans, A.; Povirk, L.; Ryan, J.J.; Kelland, L.; Farrell, N.P. Synthesis, characterization, and cytotoxicity of a novel highly charged trinuclear platinum compound. Enhancement of cellular uptake with charge. Inorg. Chem 2005, 44, 9598–9600. [Google Scholar]
- Kapp, T.; Muller, S.; Gust, R. Dinuclear alkylamine platinum (II) complexes of [1,2-bis (4-fluorophenyl) ethylenediamine] platinum (II): Influence of endocytosis and copper and organic cation transport systems on cellular uptake. ChemMedChem 2006, 1, 560–564. [Google Scholar]
- Mangrum, J.B.; Farrell, N.P. Excursions in polynuclear platinum DNA binding. Chem. Commun 2010, 46, 6640–6650. [Google Scholar]
- Montero, E.I.; Benedetti, B.T.; Mangrum, J.B.; Oehlsen, M.J.; Qu, Y.; Farrell, N.P. Pre-association of polynuclear platinum anticancer agents on a protein, human serum albumin. Implications for drug design. Dalton Trans 2007, 36, 4938–4942. [Google Scholar]
- Washizu, H.; Kikuchi, K. Electric polarizability of DNA in aqueous salt solution. J. Phys. Chem. B 2006, 110, 2855–2861. [Google Scholar]
- Bao, G.; Suresh, S. Cell and molecular mechanics of biological materials. Nat. Mater 2003, 2, 715–725. [Google Scholar]
- Komeiji, Y.; Mochizuki, Y.; Nakano, T.; Mori, H. Recent advances in fragment molecular orbitalbased molecular dynamics (FMO-MD) simulations. In Molecular Dynamics-Theoretical Developments and Applications in Nanotechnology and Energy; Wang, L.C., Ed.; InTech: Winchester, UK, 2012. [Google Scholar]
- Mori, H.; Hirayama, N.; Komeiji, Y.; Mochizuki, Y. Differences in hydration between cis- and trans- platin: Quantum insights by ab initio fragment molecular orbital-based molecular dynamics (FMO-MD). Comput. Theor. Chem 2012, 986, 30–34. [Google Scholar]
- Srinivasan, J.; Cheatham, T.E.; Cieplak, P.; Kollman, P.A.; Case, D.A. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helices. J. Am. Chem. Soc 1998, 120, 9401–9409. [Google Scholar]
- Yang, D.; van Boom, S.S.G.E.; Reedijk, J.; van Boom, J.H.; Wang, A.H.J. Structure and isomerization of an intrastrand cisplatin-cross-linked octamer DNA duplex by NMR analysis. Biochemistry 1995, 34, 12912–12920. [Google Scholar]
- Spingler, B.; Whittington, D.A.; Lippard, S.J. 2.4 Å crystal structure of an oxaliplatin1,2-d (GpG) intrastrand cross-link in a DNA dodecamer duplex. Inorg. Chem 2001, 40, 5596–5602. [Google Scholar]
- Marzilli, L.G.; Saad, J.S.; Kuklenyik, Z.; Keating, K.A.; Xu, Y. Relationship of solution and protein-bound structures of DNA duplexes with the major intrastrand cross-link lesions formed on cisplatin binding to DNA. J. Am. Chem. Soc 2001, 123, 2764–2770. [Google Scholar]
- Wheate, N.J.; Collins, J.G. Multi-nuclear platinum complexes as anti-cancer drugs. Coordin. Chem. Rev 2003, 241, 133–145. [Google Scholar]
- Grant Collins, J.; Wheate, N.J. Potential adenine and minor groove binding platinum complexes. J. Inorg. Biochem 2004, 98, 1578–1584. [Google Scholar]
- Stofer, E.; Lavery, R. Measuring the geometry of DNA grooves. Biopolymers 1994, 34, 337–346. [Google Scholar]
- Poncin, M.; Piazzola, D.; Lavery, R. DNA flexibility as a function of allomorphic conformation and of base sequence. Biopolymers 1992, 32, 1077–1103. [Google Scholar]
- Goodsell, D.S.; Dickerson, R.E. Bending and curvature calculations in B-DNA. Nucleic Acids Res 1994, 22, 5497–5503. [Google Scholar]
- Case, D.A.; Darden, T.A.; Cheatham, I.T.E.; Simmerling, C.L.; Wang, J.M.; Duke, R.E.; Luo, R.; Merz, K.M.; Pearlman, D.A.; Crowley, M. AMBER9; University of California: San Francisco, CA, USA, 2006. [Google Scholar]
- Pople, J.; Nesbet, R. Self-consistent orbitals for radicals. J. Chem. Phys 1954, 22, 571–572. [Google Scholar]
- Yao, S.; Plastaras, J.P.; Marzilli, L.G. A molecular mechanics AMBER-type force field for modeling platinum complexes of guanine derivatives. Inorg. Chem 1994, 33, 6061–6077. [Google Scholar]
- McWeeny, R.; Diercksen, G. Self-consistent perturbation Theory. II. Extension to open shells. J. Chem. Phys 1968, 49, 4852–4857. [Google Scholar]
- Chval, Z.; Sip, M. Force field for platinum binding to adenine and guanine taking into account flexibility of nucleic acids bases. J. Phys. Chem. B 1998, 102, 1659–1661. [Google Scholar]
- Cundari, T.R.; Fu, W.; Moody, E.W.; Slavin, L.L.; Snyder, L.A.; Sommerer, S.O.; Klinckman, T.R. Molecular mechanics force field for platinum coordination complexes. J. Phys. Chem 1996, 100, 18057–18064. [Google Scholar]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem 2003, 24, 1999–2012. [Google Scholar]
- Lee, M.C.; Duan, Y. Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model. Proteins Struct. Funct. Bioinf 2004, 55, 620–634. [Google Scholar]
- Perez, A.; Marchan, I.; Svozil, D.; Sponer, J.; Cheatham, T.E., III; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: Improving the description of [alpha]/[gamma] conformers. Biophys. J 2007, 92, 3817–3829. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem 2004, 25, 1157–1174. [Google Scholar]
- Lavery, R.; Sklenar, H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn 1988, 6, 63–91. [Google Scholar]
- Strahs, D.; Schlick, T. A-tract bending: Insights into experimental structures by computational models. J. Mol. Biol 2000, 301, 643–663. [Google Scholar]
- Pendyala, L.; Creaven, P. In vitro cytotoxicity, protein binding, red blood cell partitioning, and biotransformation of oxaliplatin. Cancer Res 1993, 53, 5970–5976. [Google Scholar]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem 1992, 13, 952–962. [Google Scholar]
- Amadei, A.; Linssen, A.; Berendsen, H.J.C. Essential dynamics of proteins. Proteins Struct. Funct. Bioinf 1993, 17, 412–425. [Google Scholar]
- Yamaguchi, H.; van Aalten, D.M.F.; Pinak, M.; Furukawa, A.; Osman, R. Essential dynamics of DNA containing a cis.syn cyclobutane thymine dimer lesion. Nucleic Acids Res 1998, 26, 1939–1946. [Google Scholar]
- Teeter, M.M.; Case, D.A. Harmonic and quasiharmonic descriptions of crambin. J. Phys. Chem 1990, 94, 8091–8097. [Google Scholar]
- Van Aalten, D.; Findlay, J.; Amadei, A.; Berendsen, H. Essential dynamics of the cellular retinol-binding protein—Evidence for ligand-induced conformational changes. Protein Eng 1995, 8, 1129–1135. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yue, H.; Yang, B.; Wang, Y.; Chen, G. Investigations of the Binding of [Pt2(DTBPA)Cl2](II) and [Pt2(TPXA)Cl2](II) to DNA via Various Cross-Linking Modes. Int. J. Mol. Sci. 2013, 14, 19556-19586. https://doi.org/10.3390/ijms141019556
Yue H, Yang B, Wang Y, Chen G. Investigations of the Binding of [Pt2(DTBPA)Cl2](II) and [Pt2(TPXA)Cl2](II) to DNA via Various Cross-Linking Modes. International Journal of Molecular Sciences. 2013; 14(10):19556-19586. https://doi.org/10.3390/ijms141019556
Chicago/Turabian StyleYue, Hongwei, Bo Yang, Yan Wang, and Guangju Chen. 2013. "Investigations of the Binding of [Pt2(DTBPA)Cl2](II) and [Pt2(TPXA)Cl2](II) to DNA via Various Cross-Linking Modes" International Journal of Molecular Sciences 14, no. 10: 19556-19586. https://doi.org/10.3390/ijms141019556




