HKT Transporters—State of the Art
Abstract
:1. Introduction
2. HKT1 vs. HKT2—Does the Nomenclature still Hold?
3. Class I HKT Transporters—Essential Roles in Na+ Detoxification
3.1. Arabidopsis AtHKT1;1
3.2. Rice OsHKT1;x
3.3. Wheat TaHKT1;4/5
3.4. Tomato HKT1;1 and HKT1;2
4. Class II HKT Transporters—A Role for K+
4.1. OsHKT2;1
4.2. TaHKT2;1
4.3. HvHKT2;1
5. Other Class II HKT Members
5.1. OsHKT2;2
5.2. OsHKT2;2/1, a New HKT Isoform in Rice
5.3. OsHKT2;3
5.4. OsHKT2;4, a HKT Member with Unusual Transport Characteristics, Involved in Ca2+ Signaling?
6. Do HKT Transporters Isolated from Mosses and Clubmosses Form a Third Class?
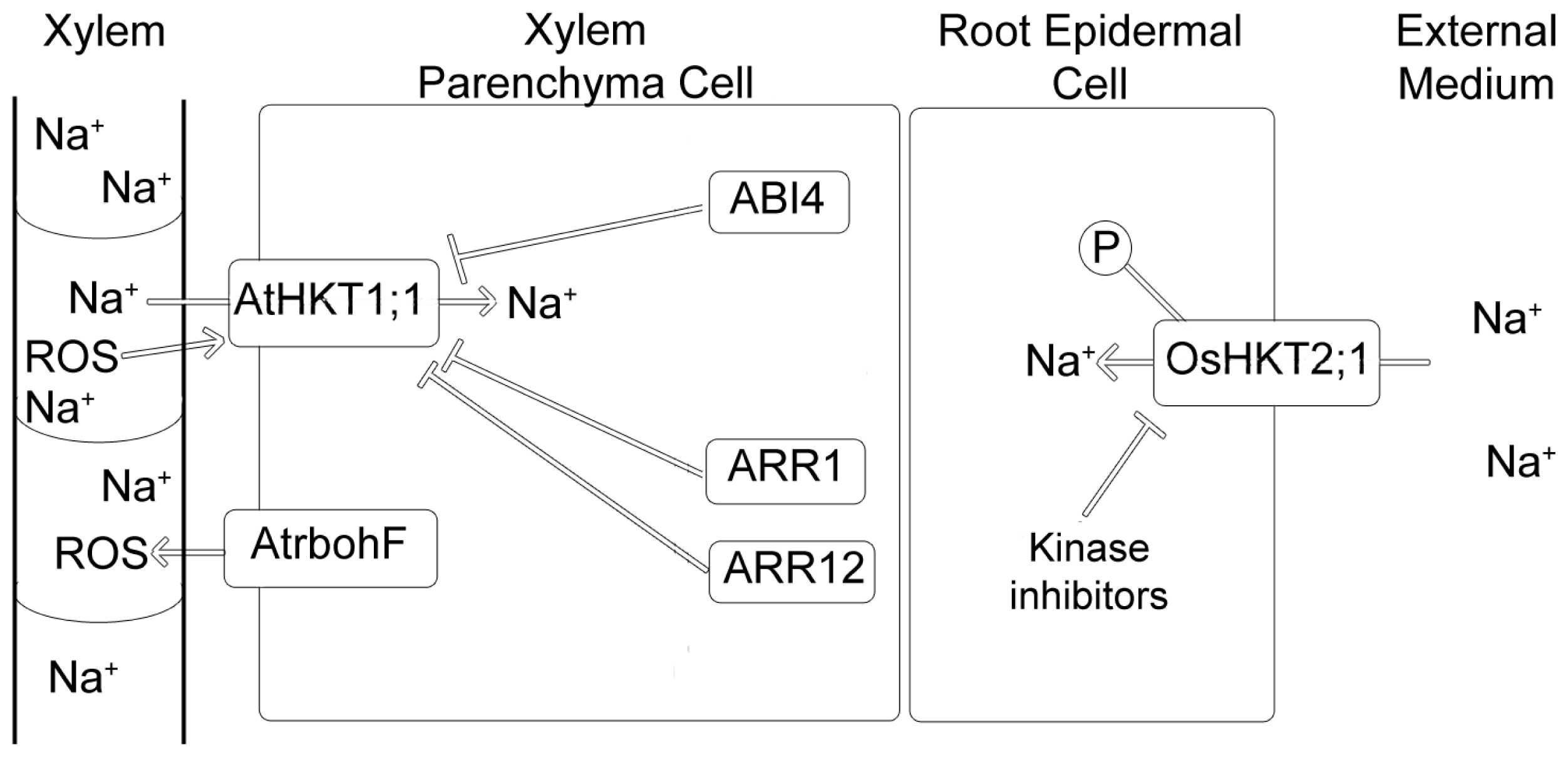
7. HKT Regulation
7.1. Promoter Structure
7.2. Regulation by ROS
7.3. Regulation by Cytokinins
7.4. Regulation by ABI4
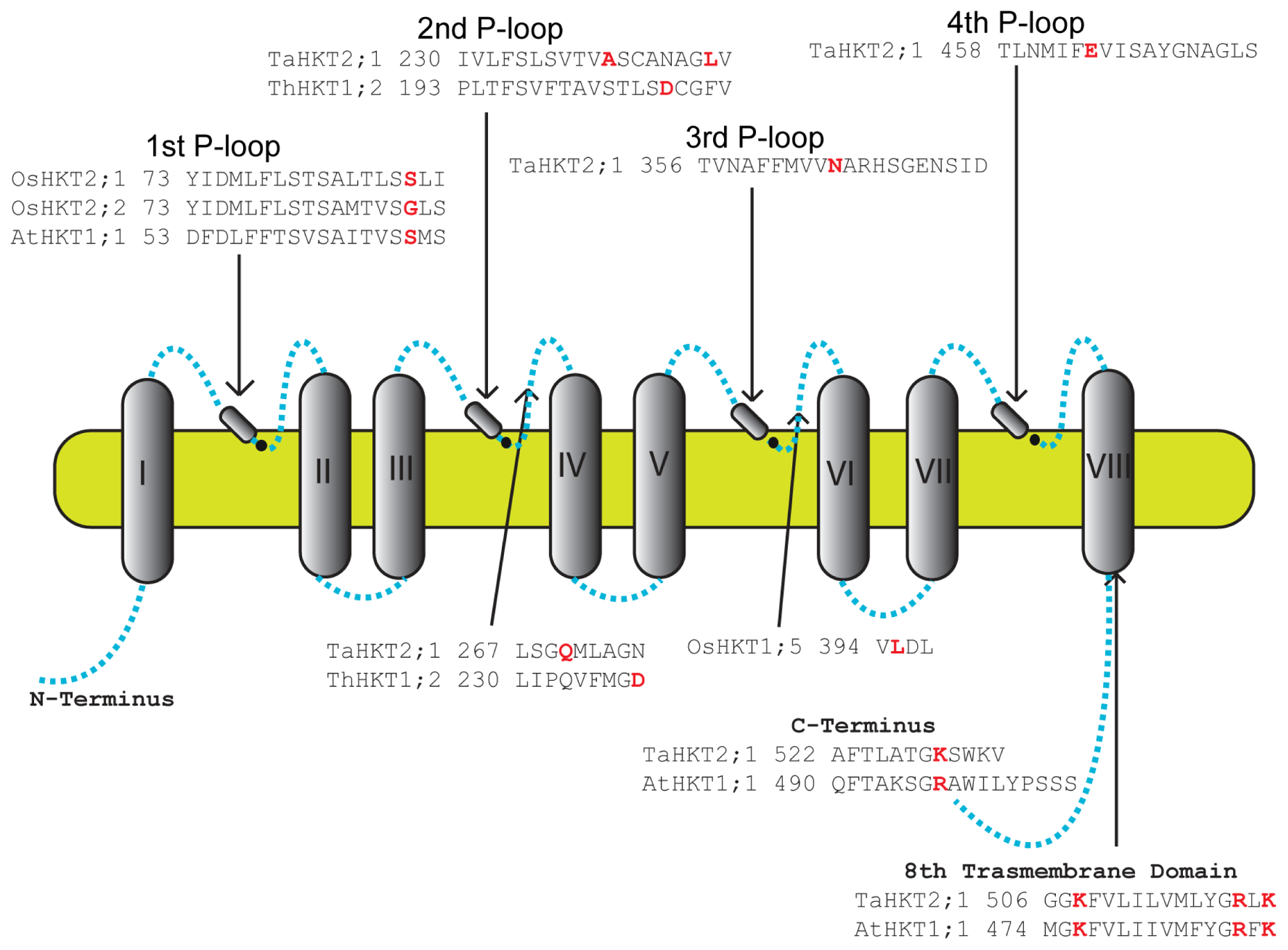
8. Residues Important for HKT Function
9. The Role in Long-Distance Transport
9.1. Recirculation vs. Exclusion: Evidence for Both Models
9.2. HKT and K+ Levels in the Xylem—A Direct or Indirect Effect of HKT
10. Future Prospects

| Transporter | Expression in planta | Ref. | Function in planta | Ref. | Transport selectivity when heterologous expressed | Ref. |
|---|---|---|---|---|---|---|
| AtHKT1;1 | Phloem (roots and shoots) | [19] | Loading excessive shoot Na+ into the phloem | [19] | Na+ transport (Xenopus oocytes) | [15] |
| Xylem parenchyma cells | [12,20,21] | Unloading of Na+ from the xylem into XPC | [12,20,21] | K+ transport (E. coli) | ||
| OsHKT1;1 | In the leaves: bulliform cells and vascular tissues. | [18,22] | Na+ transport (Xenopus oocytes and S. cerevisiae yeast cells) | [18,22] | ||
| In the roots: similar to OsHKT2;1. | ||||||
| OsHKT1;2 | Not detected in roots. | [23] | ||||
| Expression does not change with NaCl stress in the leaves. | ||||||
| OsHKT1;3 | In the leaves: bulliform cells and vascular tissues, mesophyll cells. In the roots: cortex and vascular tissues in the stele. | [22] | Na+ transport (Xenopus oocytes) | [22] | ||
| OsHKT1;4 | Leaf sheaths. | [24] | Control of sheath-to-blade transfer of Na+ | [24] | ||
| OsHKT1;5 | Mainly expressed in xylem parenchyma cells of both roots and leaves. | [10] | Control of root-to-shoot transfer of Na+ by unloading of Na+ from the xylem into XPC | [10] | Na+ transport (Xenopus oocytes) | [10] |
| TaHKT1;4 | Expressed in the roots, leaf sheath and leaf blades. | [25] | Unloading of Na+ from the xylem into XPC | [25] | ||
| TaHKT1;5 | Expressed in the roots but not in the shoots. | [7] | Unloading of Na+ from the xylem into XPC | [7,13] | Na+ transport (S. cerevisiae cell) | [13] |
| Na+ transport (Xenopus oocytes) | ||||||
| Na+ transport (S. cerevisiae cells) | [26] | |||||
| SlHKT1;1 | Ubiquitously expressed (roots, stems, leaves, flowers, fruits). | [26] | Na+ transport (Xenopus oocytes) | Almeida et al. unpublished results | ||
| NIL and treatment dependent | ||||||
| SlHKT1;2 | Ubiquitously expressed (roots, stems, leaves, flowers, fruits). NIL and treatment dependent | [26] | No transport activity detected in either S. cerevisiae cells or Xenopus oocytes | [26] | ||
| Almeida et al. unpublished results | ||||||
| EcHKT1;1 | Expressed in the leaves, stems and roots | [27] | K+ transport (E. coli cells) | [27,28] | ||
| Na+, K+ and Rb+ transport (Xenopus oocytes) | ||||||
| EcHKT1;2 | Expressed in the leaves, stems and roots | [27] | K+ transport (E. coli cells) | [27,28] | ||
| Na+, K+, Rb+, Li+ Transport (Xenopus oocytes) | ||||||
| McHKT1;1 | In the leaves: xylem parenchyma cells and phloem cells; In the roots: epidermal cells and vascular tissues | [29] | The authors proposed a model where McHKT1;1 Unloads Na+ from the xylem in the shoots | [29] | K+ transport (S. cerevisiae cells) | [29] |
| Rb+, Cs+, K+, Na+ and Li+ transport (Xenopus oocytes) |
| Transporter | Expression in planta | Ref. | Function in planta | Ref. | Transport selectivity when heterologous expressed | Ref. |
|---|---|---|---|---|---|---|
| OsHKT2;1 | In the roots: epidermis, exodermis, cortex differentiated into aerenchyma, stele (mainly in the phloem); In the leaves: bulliform cells, xylem, phloem, mesophyll cells | [22] | Nutritional Na+ uptake from the external medium | [11] | Na+ and K+ transport (Xenopus oocytes) | [22,30,31] |
| Na+ transport (S. cerevisiae cells) | [18,30] | |||||
| K+ transport (S. cerevisiae cells) | [31] | |||||
| OsHKT2;2 | Expressed only in the roots | [32] | Na+/K+ symporter in BY2 tobacco cells | [33] | Na+, K+, (S. cerevisiae and Xenopus oocytes) | [30,34] |
| Expected to co-transport both Na+ and K+ in conditions of K+ starvation | [32] | |||||
| OsHKT2;2/1 | Expressed only in the roots | [32] | Expected to co-transport both Na+ and K+ in the roots in conditions of low K+ and under salt stress | [32] | Na+ and K+ transport (S. cerevisiae cells and Xenopus oocytes) | [32] |
| OsHKT2;3 | Marginally expressed in the roots in comparison to the shoots | [34] | No currents or uptake observed in Xenopus oocytes or S. cerevisiae cells | [34] | ||
| OsHKT2;4 | Vasculature of primary/ lateral root cells, leaf sheaths, spikelets and the base of stems. Expressed also in mesophyll cells | [35] | Possible role in K+ homeostasis as a K+ transporter/channel | [34] | Permeable to NH4+, Li+, Na+, K+, Ca2+, Mg2+ Zn2+, Mn2+, Cu2+, Fe2+, Cd2+ (Xenopus oocytes) | [35] |
| Possible redundant role in planta as oshkt2;4 mutants show no phenotype | [35] | Permeable to Na+, K+, Mg2+, Ca2+ (Xenopus oocytes) | [34] | |||
| K+ transport (S. cerevisiae cells) | ||||||
| Proposed to function as a K+ transporter involved in both nutritional K+ uptake and long-distance K+ transport | [36] | Na+ and K+ transport (Xenopus oocytes) | [36] | |||
| TaHKT2;1 | Root cortical and stele cells Vascular tissue of mesophyll cells | [14] | Na+ uptake from the external medium | [37] | Permeable to Na+, K+, Cs+ and Rb+ (Xenopus oocytes) | [14] |
| K+ transport (S. cerevisiae cells) | ||||||
| Na+ and K+ transport (S. cerevisiae cells) | [38,39] | |||||
| Na+ and K+ transport (Xenopus oocytes) | ||||||
| Permeable to Na+, K+ and Mg+ (Xenopus oocytes) | [34] | |||||
| PutHKT2;1 | Mainly in roots | [40] | Possible high affinity K+ transporter | [40] | Na+ and K+ transport (S. cerevisiae cells) | [40] |
| HvHKT2;1 | Root cortex, leaf blades and leaf sheaths | [41,42] | Possible involvement in the root K+ (re)absorption at very low K+ concentrations | [41,42] | Na+ and K+ transporter (S. cerevisiae and Xenopus oocytes) | [41–43] |
| Possible uptake of Na+ in the roots | ||||||
| PhaHKT2;1 | Roots and shoots | [44] | Na+ and K+ transport (S. cerevisiae cells) | [40,44] | ||
| PpHKT1 | ------- | ------- | Na+ and K+ uptake (S. cerevisiae cells) | [45] |
Conflicts of Interest
References
- Huang, S.; Spielmeyer, W.; Lagudah, E.S.; Munns, R. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J. Exp. Bot 2008, 59, 927–937. [Google Scholar]
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot 2006, 57, 1017–1023. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol 2008, 59, 651–681. [Google Scholar]
- Shi, H.; Ishitani, M.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar]
- Shi, H.Z.; Lee, B.H.; Wu, S.J.; Zhu, J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol 2003, 21, 81–85. [Google Scholar]
- Maser, P.; Eckelman, B.; Vaidyanathan, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimura, M.; Uozumi, N.; et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT 1;1. FEBS Lett 2002, 531, 157–161. [Google Scholar]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.; Munns, R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 2007, 143, 1918–1928. [Google Scholar]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci 2009, 14, 660–668. [Google Scholar]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet 2005, 37, 1141–1146. [Google Scholar]
- Horie, T.; Costa, A.; Kim, T.H.; Han, M.J.; Horie, R.; Leung, H.Y.; Miyao, A.; Hirochika, H.; An, G.; Schroeder, J.I. Rice OsHKT2;1 transporter mediates large Na+ influx components into K+-starved roots for growth. EMBO J 2007, 26, 3003–3014. [Google Scholar]
- Moller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.J.; Coates, J.C.; Haseloff, J.; Tester, M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, J.S.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol 2012, 30, 360–364. [Google Scholar]
- Schachtman, D.P.; Schroeder, J.I. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher-plants. Nature 1994, 370, 655–658. [Google Scholar]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsuboi, A.; Bakker, E.P.; Nakamura, T.; Schroeder, J.I. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 2000, 122, 1249–1259. [Google Scholar]
- Platten, J.D.; Cotsaftis, O.; Berthomieu, P.; Bohnert, H.; Davenport, R.J.; Fairbairn, D.J.; Horie, T.; Leigh, R.A.; Lin, H.X.; Luan, S.; et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 2006, 11, 372–374. [Google Scholar]
- Maser, P.; Hosoo, Y.; Goshima, S.; Horie, T.; Eckelman, B.; Yamada, K.; Yoshida, K.; Bakker, E.P.; Shinmyo, A.; Oiki, S.; et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc. Nat. Acad. Sci. USA 2002, 99, 6428–6433. [Google Scholar]
- Garciadeblas, B.; Senn, M.E.; Banuelos, M.A.; Rodriguez-Navarro, A. Sodium transport and HKT transporters: The rice model. Plant J 2003, 34, 788–801. [Google Scholar]
- Berthomieu, P.; Conejero, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F.; et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 2003, 22, 2004–2014. [Google Scholar]
- Plett, D.; Safwat, G.; Gilliham, M.; Moller, I.S.; Roy, S.; Shirley, N.; Jacobs, A.; Johnson, A.; Tester, M. Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS One 2010, 5, e12571. [Google Scholar]
- Sunarpi; Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.Y.; Leung, H.Y.; Hattori, K.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J 2005, 44, 928–938. [Google Scholar]
- Jabnoune, M.; Espeout, S.; Mieulet, D.; Fizames, C.; Verdeil, J.L.; Conejero, G.; Rodriguez-Navarro, A.; Sentenac, H.; Guiderdoni, E.; Abdelly, C.; et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol 2009, 150, 1955–1971. [Google Scholar]
- Wu, Y.S.; Hu, Y.B.; Xu, G.H. Interactive effects of potassium and sodium on root growth and expression of K+/Na+ transporter genes in rice. Plant Growth Regul 2009, 57, 271–280. [Google Scholar]
- Cotsaftis, O.; Plett, D.; Shirley, N.; Tester, M.; Hrmova, M. A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS One 2012, 7, e39865. [Google Scholar]
- Huang, S.B.; Spielmeyer, W.; Lagudah, E.S.; James, R.A.; Platten, J.D.; Dennis, E.S.; Munns, R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 2006, 142, 1718–1727. [Google Scholar]
- Asins, J.M.; Villalta, I.; Aly, M.M.; Olias, R.; Morales, P.A.; Huertas, R.; Li, J.; Jaime-Perez, N.; Haro, R.; Raga, V.; et al. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ 2012, 36, 1171–1191. [Google Scholar]
- Fairbairn, D.J.; Liu, W.H.; Schachtman, D.P.; Gomez-Gallego, S.; Day, S.R.; Teasdale, R.D. Characterisation of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis. Plant Mol. Biol 2000, 43, 515–525. [Google Scholar]
- Liu, W.; Fairbairn, D.J.; Reid, R.J.; Schachtman, D.P. Characterization of two HKT1 homologues from Eucalyptus camaldulensis that display intrinsic osmosensing capability. Plant Physiol 2001, 127, 283–294. [Google Scholar]
- Su, H.; Balderas, E.; Vera-Estrella, R.; Golldack, D.; Quigley, F.; Zhao, C.S.; Pantoja, O.; Bohnert, J.H. Expression of the cation transporter McHKT1 in a halophyte. Plant Mol. Biol 2003, 52, 967–980. [Google Scholar]
- Horie, T.; Yoshida, K.; Nakayama, H.; Yamada, K.; Oiki, S.; Shinmyo, A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 2001, 27, 129–138. [Google Scholar]
- Golldack, D.; Su, H.; Quigley, F.; Kamasani, U.R.; Munoz-Garay, C.; Balderas, E.; Popova, O.V.; Bennett, J.; Bohnert, H.J.; Pantoja, O. Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J 2002, 31, 529–542. [Google Scholar]
- Oomen, R.J.; Benito, B.; Sentenac, H.; rodriguez-Navarro, A.; Talon, M.; Very, A.A.; Domingo, C. HKT2;2/1, a K+ permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J 2012, 71, 750–762. [Google Scholar]
- Yao, X.; Horie, T.; Xue, S.W.; Leung, H.Y.; Katsuhara, M.; Brodsky, D.E.; Wu, Y.; Schroeder, J.I. Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol 2010, 152, 341–355. [Google Scholar]
- Horie, T.; Brodsky, D.E.; Costa, A.; Kaneko, T.; Lo Schiavo, F.; Katsuhara, M.; Schroeder, J.I. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol 2011, 156, 1493–1507. [Google Scholar]
- Lan, W.Z.; Wang, W.; Wang, S.M.; Li, L.G.; Buchanan, B.B.; Lin, H.X.; Gao, J.P.; Luan, S. A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proc. Natl. Acad. Sci. USA 2010, 107, 7089–7094. [Google Scholar]
- Sassi, A.; Meieulet, D.; Khan, I.; Moreau, B.; Gaillard, I.; Sentenac, H.; Very, A.A. The rice monovalent cation transporter OsHKT2;4: Revisited ionic selectivity. Plant Physiol 2012, 160, 498–510. [Google Scholar]
- Laurie, S.; Feeney, K.A.; Maathuis, F.J.M.; Heard, P.J.; Brown, S.J.; Leigh, R.A. A role for HKT1 in sodium uptake by wheat roots. Plant J 2002, 32, 139–149. [Google Scholar]
- Rubio, F.; Schwarz, M.; Gassmann, W.; Schroeder, J.I. Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. J. Biol. Chem 1999, 12;274, 6839–6847. [Google Scholar]
- Rubio, F.; Gassmann, W.; Schroeder, J.I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 1995, 270, 1660–1663. [Google Scholar]
- Ardie, S.W.; Xie, L.; Takahashi, R.; Liu, S.; Takano, T. Cloning of a high-affinity K+ transporter gene PutHKT2;1 from Puccinellia tenuiflora and its functional comparison with OsHKT2;1 from rice in yeast and Arabidopsis. J. Exp. Bot 2009, 60, 3491–3502. [Google Scholar]
- Haro, R.; Banuelos, M.A.; Senn, M.A.E.; Barrero-Gil, J.; Rodriguez-Navarro, A. HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol 2005, 139, 1495–1506. [Google Scholar]
- Mian, A.; Oomen, R.J.; Isayenkow, S.; Sentenac, H.; Maathuis, F.J.; Very, A.A. Overexpression of a Na+ and K+-permeable HKT transporter in barley improves salt tolerance. Plant J 2011, 68, 468–479. [Google Scholar]
- Banuelos, M.A.; Haro, R.; Fraile-Escanciano, A.; Rodriguez-Navarro, A. Effects of polylinker uATGs on the function of grass HKT1 transporters expressed in yeast cells. Plant Cell Physiol 2008, 49, 1128–1132. [Google Scholar]
- Takahashi, R.; Liu, S.; Takano, T. Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and salt-sensitive reed plants. J. Exp. Bot 2007, 58, 4387–4395. [Google Scholar]
- Moller, I.S.; Tester, M. Salinity tolerance of Arabidopsis: A good model for cereals? Trends Plant Sci 2007, 12, 534–540. [Google Scholar]
- Ali, Z.; Park, H.C.; Ali, A.; Oh, D.-H.; Aman, R.; Kropornicka, A.; Hong, H.; Choi, W.; Chung, S.S.; Kim, W.-Y.; et al. TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol 2012, 158, 1463–1474. [Google Scholar]
- Rus, A.; Yokoi, S.; Sharkhuu, A.; Reddy, M.; Lee, B.H.; Matsumoto, T.K.; Koiwa, H.; Zhu, J.K.; Bressan, R.A.; Hasegawa, P.M. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. USA 2001, 98, 14150–14155. [Google Scholar]
- Essah, P.A.; Davenport, R.; Tester, M. Sodium influx and accumulation in Arabidopsis. Plant Physiol 2003, 133, 307–318. [Google Scholar]
- Davenport, R.J.; Munoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 2007, 30, 497–507. [Google Scholar]
- Haseloff, J. GFP variants for multispectral imaging of living cells. Method Cell Biol 1999, 58, 139–151. [Google Scholar]
- Rus, A.; Lee, B.H.; Munoz-Mayor, A.; Sharkhuu, A.; Miura, K.; Zhu, J.K.; Bressan, R.A.; Hasegawa, P.M. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 2004, 136, 2500–2511. [Google Scholar]
- Munns, R. Salinity, Growth and Phytohormones. In Salinity: Environment-Plants-Molecules; Lauchli, A., Luttge, U., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; pp. 271–290. [Google Scholar]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot 2003, 91, 503–527. [Google Scholar]
- Munns, R. Physiological processes limiting plant growth in saline soils: Some dogmas and hypothesis. Plant Cell Environ 1993, 16, 15–24. [Google Scholar]
- Rus, A.; Baxter, I.; Muthukumar, B.; Gustin, J.; Lahner, B.; Yakubova, E.; Salt, D.E. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2006, 2, 1964–1973. [Google Scholar]
- Jha, D.; Shirley, N.; Tester, M.; Roy, S.J. Variation in salinity tolerance and shoot sodium accumulation in Arabidopsis ecotypes linked to differences in the natural expression levels of transporters involved in sodium transport. Plant Cell Environ 2010, 33, 793–804. [Google Scholar]
- Gorham, J.; Bristol, A.; Young, E.M.; Jones, R.G.W.; Kashour, G. Salt tolerance in the triticeae—K+/Na+ discrimination in barley. J. Exp. Bot 1990, 41, 1095–1101. [Google Scholar]
- Dubcovsky, J.; Santa-Maria, G.; Epstein, E.; Luo, M.C.; Dvoak, J. Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor. Appl. Genet 1996, 92, 448–454. [Google Scholar]
- Gorham, J.; Hardy, C.; Wyn, J.R.G.; Joppa, L.R.; Law, C.N. Chromosomal location of a K+/Na+ discrimination character in the D genome of wheat. Theor. Appl. Genet 1987, 74, 584–588. [Google Scholar]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot 2011, 62, 2939–2947. [Google Scholar]
- James, R.A.; Davenport, R.J.; Munns, R. Physiological characterisation of two genes for Na+ exclusion in durum wheat: Nax1 and Nax2. Plant Physiol 2006, 142, 1537–1547. [Google Scholar]
- Plett, D.C.; Moller, I.S. Na+ transport in glycophytic plants: What we know and would like to know. Plant Cell Environ 2010, 33, 612–626. [Google Scholar]
- Haro, R.; Banuelos, M.A.; Rodriguez-Navarro, A. High-affinity sodium uptake in land plants. Plant Cell Physiol 2010, 51, 68–79. [Google Scholar]
- Horie, T.; Sugawara, M.; Okunou, K.; Nakayama, H.; Schroeder, J.I.; Shinmyo, A.; Yoshida, K. Functions of HKT transporters in sodium transport in roots and in protecting leaves from salinity stress. Plant Biotechnol 2008, 25, 233–239. [Google Scholar]
- Rodriguez-Navarro, A.; Rubio, F. High-affinity potassium and sodium transport systems in plants. J. Exp. Bot 2006, 57, 1149–1160. [Google Scholar]
- Wang, T.B.; Gassmann, W.; Rubio, F.; Schroeder, J.I.; Glass, A.D.M. Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol 1998, 118, 651–659. [Google Scholar]
- Buschmann, P.H.; Vaidyanathan, R.; Gassmann, W.; Schroeder, J.I. Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol 2000, 122, 1387–1397. [Google Scholar]
- Maathuis, F.J.M.; Ichida, A.M.; Sanders, D.; Schroeder, J.I. Roles of higher plant K+ channels. Plant Physiol 1997, 114, 1141–1149. [Google Scholar]
- Cao, Y.; Jin, X.S.; Huang, H.; Derebe, M.G.; Levin, E.J.; Kabaleeswaran, V.; Pan, Y.P.; Punta, M.; Love, J.; Weng, J.; et al. Crystal structure of a potassium ion transporter, TrkH. Nature 2011, 471, 336–340. [Google Scholar]
- Durell, S.R.; Guy, H.R. Structural models of the KtrB, TrkH, and Trk1,2 symporters based on the structure of the KcsA K+ channel. Biophys. J 1999, 77, 789–807. [Google Scholar]
- Durell, S.R.; Hao, Y.L.; Nakamura, T.; Bakker, E.P.; Guy, H.R. Evolutionary relationship between K+ channels and symporters. Biophys. J 1999, 77, 775–788. [Google Scholar]
- Gassmann, W.; Rubio, F.; Schroeder, J.I. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J 1996, 10, 869–882. [Google Scholar]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 2010, 33, 552–565. [Google Scholar]
- Baek, D.; Jiang, J.F.; Chung, J.S.; Wang, B.S.; Chen, J.P.; Xin, Z.G.; Shi, H.Z. Regulated AtHKT1;1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol 2011, 52, 149–161. [Google Scholar]
- Miller, G.; Suzuki, N.; Rizhsky, L.; Hegie, A.; Koussevitzky, S.; Miller, R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol 2007, 144, 1777–1785. [Google Scholar]
- Kaye, Y.; Golani, Y.; Singer, Y.; Leshen, Y.; Cohen, G.; Ercetin, M.; Gillaspy, G.; Levine, A. Inositol polyphosphate 5-phosphatase7 regulates production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol 2011, 157, 229–241. [Google Scholar]
- Leshem, Y.; Seri, L.; Levine, A. Induction of phosphotidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J 2007, 51, 185–197. [Google Scholar]
- Jiang, C.; Belfield, E.J.; Mithani, A.; Visscher, A.; Ragoussis, J.; Mott, R.; Smith, J.A.C.; Harberd, N.P. Ros-mediated vascular homeostatic control of root-to-shoot soil Na+ delivery in Arabidopsis. EMBO J 2012, 31, 4359–4370. [Google Scholar]
- Garcia-Mata, C.; Wang, J.; Gajdanowicz, P.; Gonzalez, W.; Hills, A.; Donald, N.; Riedelsberger, J.; Amtmann, A.; Dreyer, I.; Blatt, M.R. A minimal cysteine motif required to activate the SKOR K+ channel of Arabidopsis by the reactive oxygen species H2O2. J. Biol. Chem 2010, 285, 29286–29294. [Google Scholar]
- Itai, C.; Richmond, A.; Vaadia, Y. The role of root cytokinins during water and salinity stress. Isr. J. Bot 17, 195.
- Mason, M.G.; Jha, D.; Salt, D.E.; Tester, M.; Hill, K.; Kieber, J.J.; Schaller, G.E. Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J 2010, 64, 753–763. [Google Scholar]
- Argueso, C.T.; Ferreira, F.J.; Kieber, J.J. Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ 2009, 32, 1147–1160. [Google Scholar]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar]
- Nishiyama, R.; Le, T.D.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Transcriptome analysis of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One 2012, 7, e32124. [Google Scholar]
- Mason, M.G.; Li, J.; Mathews, D.E.; Kieber, J.J.; Schaller, G.E. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 2004, 135, 927–937. [Google Scholar]
- Zhang, H.; Kim, M.S.; Sun, Y.; Dowd, S.E.; Shi, H.Z.; Pare, P.W. Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant-Microbe Interact 2008, 21, 737–744. [Google Scholar]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Wei, H.X.; Pare, P.W.; Kloepper, J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 4927–4932. [Google Scholar]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S. ABI4: Versatile activator and repressor. Trends Plant Sci 2013, 18, 125–132. [Google Scholar]
- Shkolnik-Inbar, D.; Adler, G.; Bar-Zvi, D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J 2013, 73, 993–1005. [Google Scholar]
- Diatloff, E.; Kumar, R.; Schachtman, D.P. Site directed mutagenesis reduces the Na+ affinity of HKT1, an Na+ energized high affinity K+ transporter. FEBS Lett 1998, 432, 31–36. [Google Scholar]
- Kato, N.; Akai, M.; Zulkifli, L.; Matsuda, N.; Kato, Y.; Goshima, S.; Hazama, A.; Yamagami, M.; Guy, H.R.; Uozumi, N. Role of positively charged amino acids in the M2D transmembrane helix of Ktr/Trk/HKT type cation transporters. Channels 2007, 1, 161–171. [Google Scholar]
- Pardo, J.M. Biotechnology of water and salinity stress tolerance. Curr. Opin. Biotech 2010, 21, 185–196. [Google Scholar] [Green Version]
- Takahashi, R.; Nishio, T.; Ichizen, N.; Takano, T. High-affinity K+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows Na+ permeability under NaCl stress. Plant Cell Rep 2007, 26, 1673–1679. [Google Scholar]
- Xue, S.; Yao, X.; Luo, W.; Jha, D.; Tester, M.; Horie, T.; Schroeder, J.I. AtHKT1;1 mediates Nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS One 2012, 6, e24725. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Almeida, P.; Katschnig, D.; De Boer, A.H. HKT Transporters—State of the Art. Int. J. Mol. Sci. 2013, 14, 20359-20385. https://doi.org/10.3390/ijms141020359
Almeida P, Katschnig D, De Boer AH. HKT Transporters—State of the Art. International Journal of Molecular Sciences. 2013; 14(10):20359-20385. https://doi.org/10.3390/ijms141020359
Chicago/Turabian StyleAlmeida, Pedro, Diana Katschnig, and Albertus H. De Boer. 2013. "HKT Transporters—State of the Art" International Journal of Molecular Sciences 14, no. 10: 20359-20385. https://doi.org/10.3390/ijms141020359




