Genetic Diversity and Population Differentiation of Calanthe tsoongiana, a Rare and Endemic Orchid in China
Abstract
:1. Introduction
2. Results
2.1. Genetic Diversity of C. tsoongiana
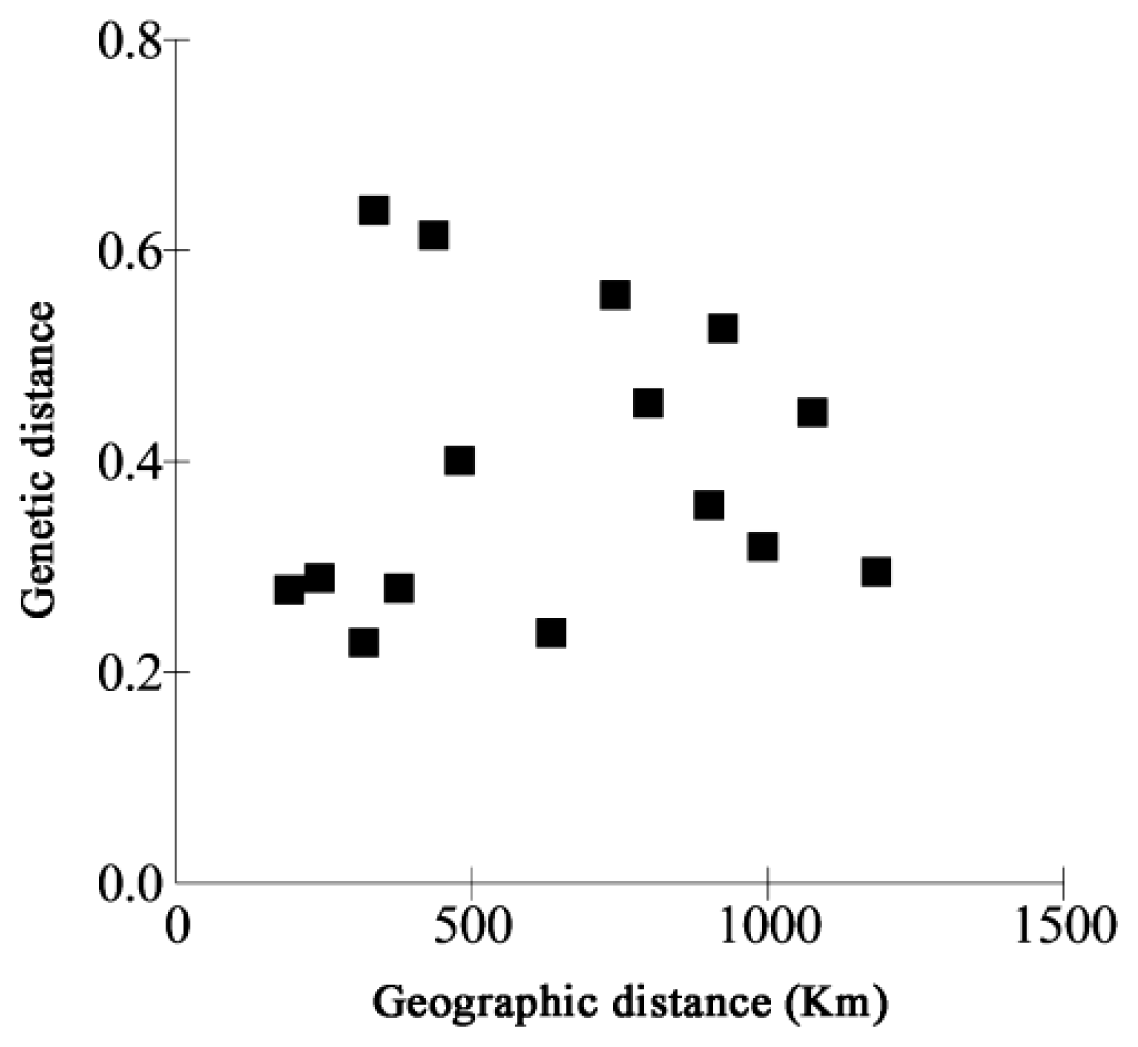
2.2. Genetic Differentiation and Gene Flow in C. tsoongiana
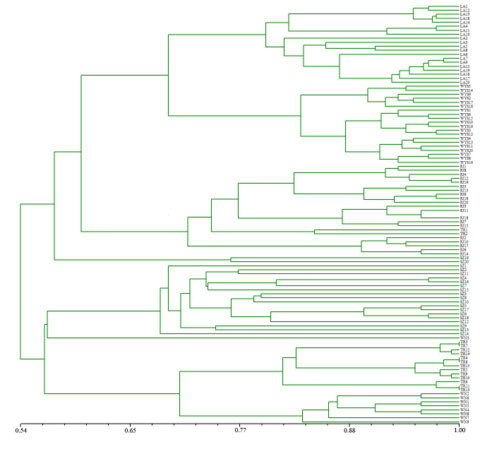
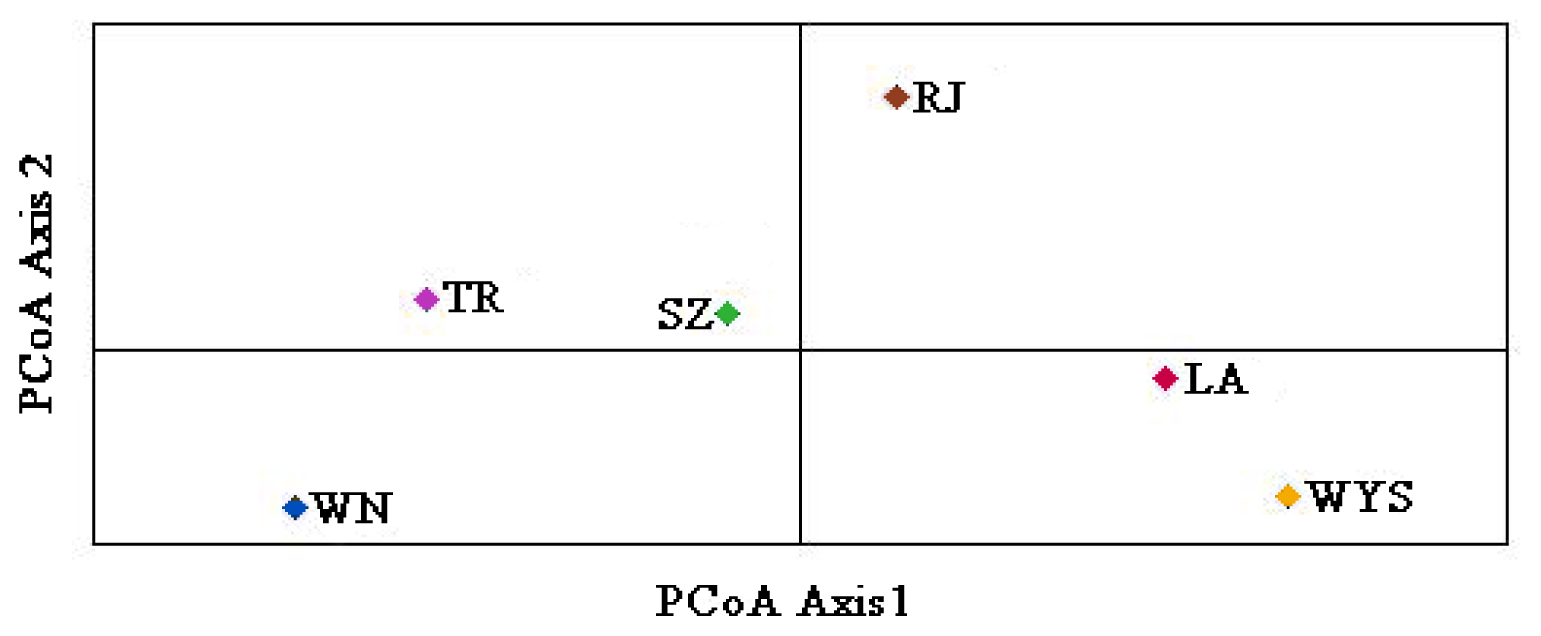
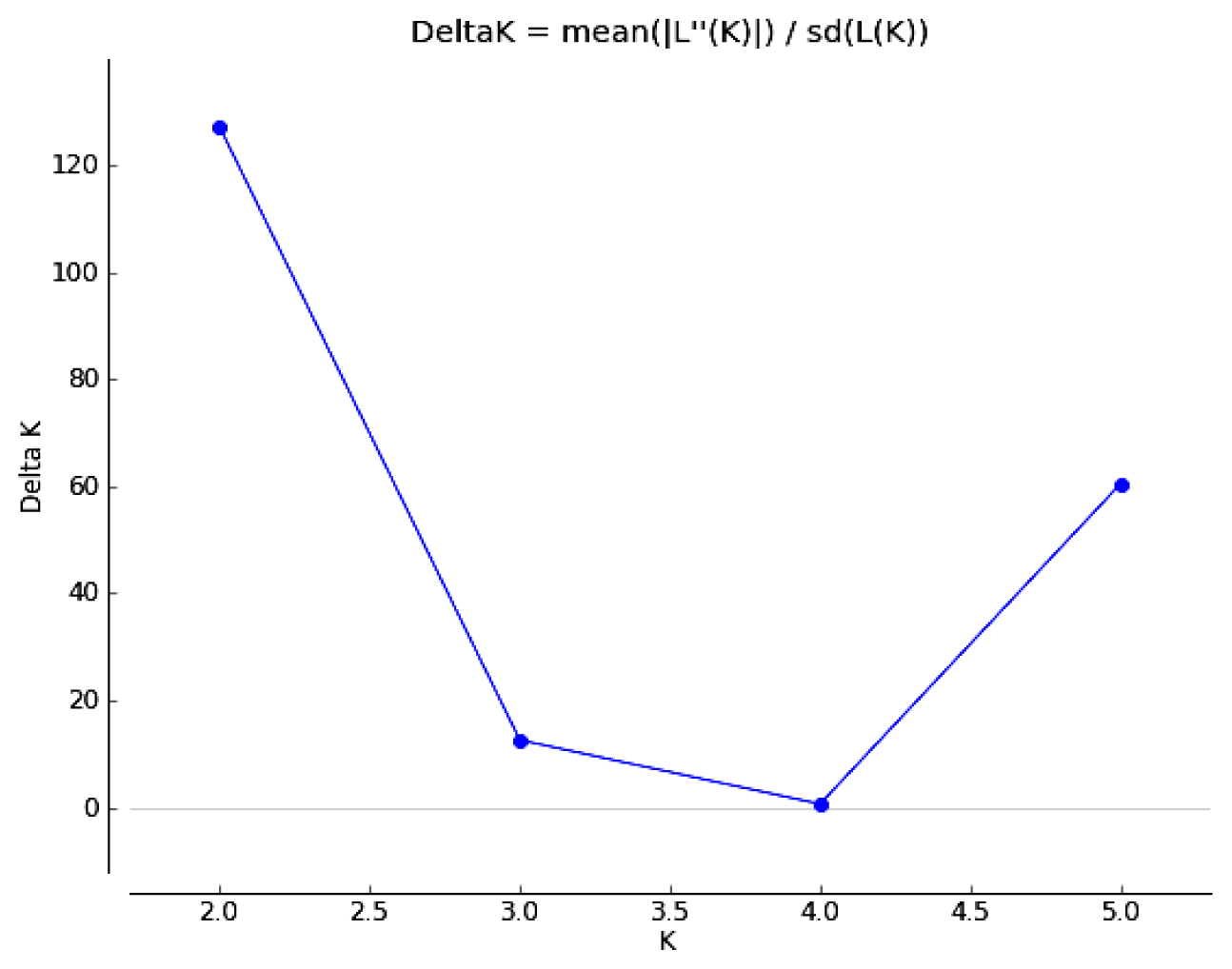
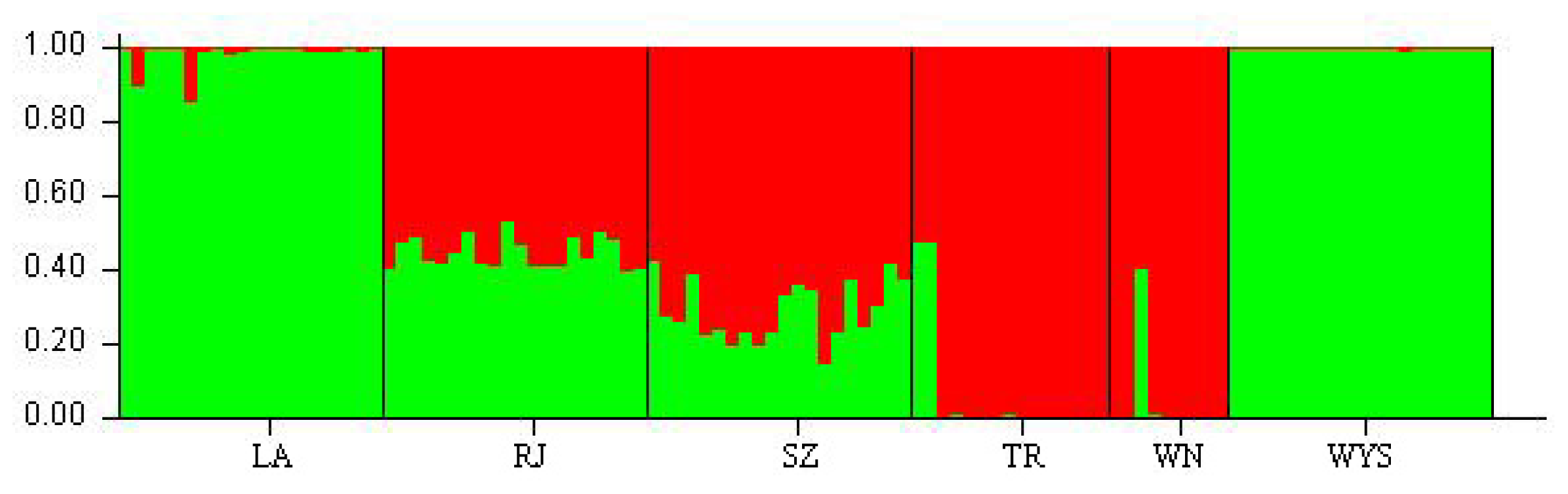
2.3. Genetic Relationships among C. tsoongiana Populations
3. Discussion
3.1. Genetic Diversity
3.2. Genetic Differentiation
3.3. Implications for Conservation
4. Materials and Methods
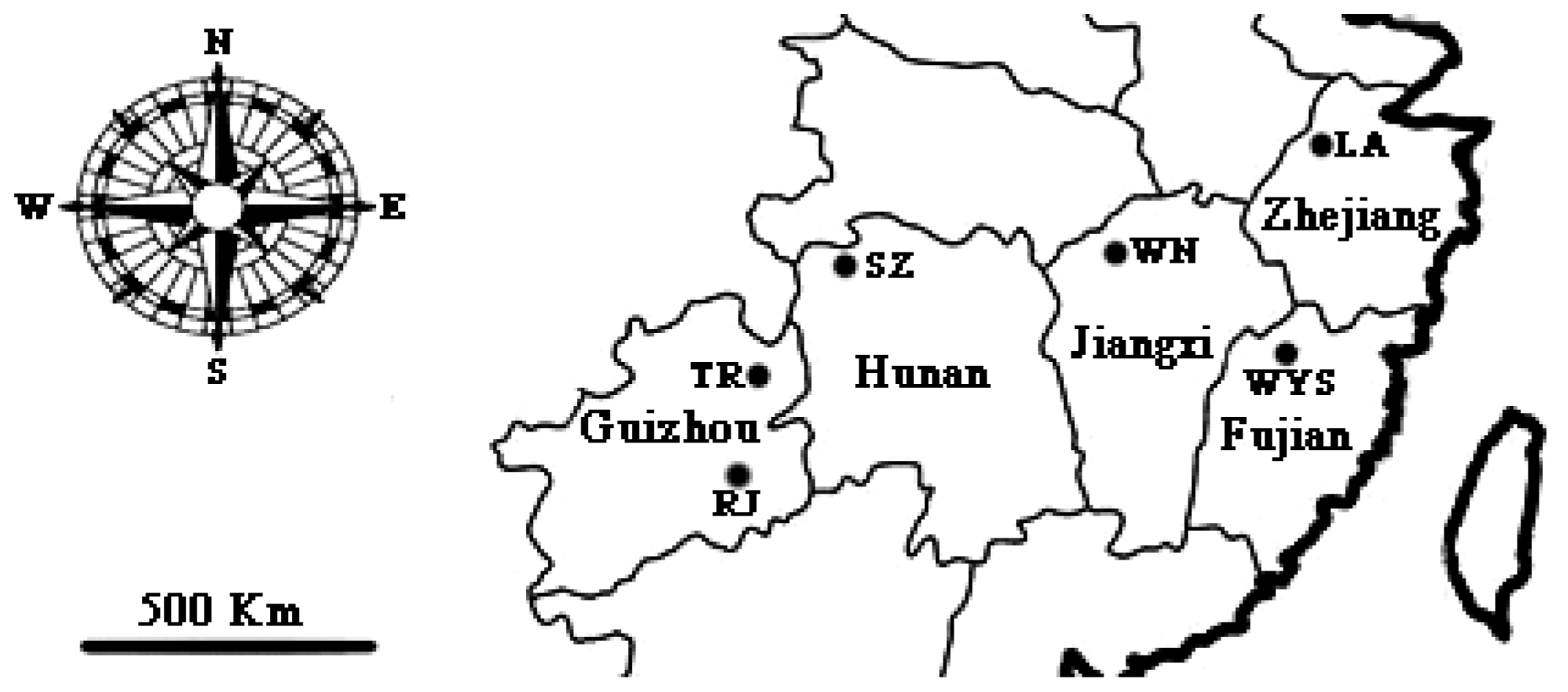
4.1. Sample Collection
4.2. DNA Extraction and PCR Amplification
4.3. Data Analysis
5. Conclusions
| Populations | Na | Ne | H | I | PPL |
|---|---|---|---|---|---|
| LA | 1.419 (0.496) | 1.286 (0.378) | 0.163 (0.206) | 0.239 (0.296) | 41.9% |
| RJ | 1.524 (0.501) | 1.314 (0.377) | 0.182 (0.200) | 0.273 (0.287) | 52.4% |
| SZ | 1.726 (0.448) | 1.504 (0.389) | 0.284 (0.201) | 0.415 (0.282) | 72.6% |
| TR | 1.694 (0.463) | 1.379 (0.360) | 0.226 (0.192) | 0.342 (0.271) | 69.4% |
| WN | 1.307 (0.463) | 1.210 (0.356) | 0.119 (0.191) | 0.174 (0.274) | 30.7% |
| WYS | 1.331 (0.472) | 1.215 (0.351) | 0.122 (0.191) | 0.180 (0.275) | 33.1% |
| Average | 1.500 (0.474) | 1.318 (0.369) | 0.183 (0.197) | 0.271 (0.281) | 50.0% |
| Species Level | 1.968 (0.177) | 1.720 (0.285) | 0.398 (0.129) | 0.576 (0.164) | 96.8% |
| SV | d.f. | SSD | MSD | VC | TVP | Φ pt | p (rand ≥ data) |
|---|---|---|---|---|---|---|---|
| Among populations | 5 | 1252.535 | 250.507 | 13.878 | 52% | 0.522 | 0.001 |
| Within populations | 98 | 1246.456 | 12.719 | 12.719 | 48% | ||
| Total | 103 | 2498.991 | 26.597 | 100% |
| Populations | LA | RJ | SZ | TR | WN | WYS |
|---|---|---|---|---|---|---|
| LA | - | 0.744 | 0.699 | 0.640 | 0.541 | 0.796 |
| RJ | 0.296 | - | 0.756 | 0.757 | 0.572 | 0.727 |
| SZ | 0.359 | 0.280 | - | 0.749 | 0.670 | 0.634 |
| TR | 0.447 | 0.279 | 0.290 | - | 0.789 | 0.591 |
| WN | 0.615 | 0.558 | 0.401 | 0.237 | - | 0.528 |
| WYS | 0.229 | 0.319 | 0.456 | 0.527 | 0.638 | - |
| Orchid Species | AM | NP | NI | NL | Gst (Fst) | SR |
|---|---|---|---|---|---|---|
| Piperia yadonii (2006) | ISSR | 7 | 210 | 636 | 0.42 | [7] |
| Piperia yadonii (2007) | ISSR | 8 | 251 | 622 | 0.39 | [7] |
| Cymbidium goeringii | ISSR | 11 | 325 | 127 | 0.22 | [11] |
| Tipularia discolor | ISSR | 4 | 56 | 22 | 0.42 | [12] |
| Platanthera aquilonis | ISSR | 7 | 111 | 164 | 0.70 | [13] |
| Platanthera dilatata | ISSR | 10 | 152 | 339 | 0.49 | [13] |
| Platanthera huronensis | ISSR | 14 | 236 | 517 | 0.36 | [13] |
| Gastrodia elata | ISSR | 14 | 483 | 77 | 0.27 | [14] |
| Dendrobium fimbriatum | ISSR | 5 | 114 | 117 | 0.75 | [15] |
| Calanthe tsoongiana | ISSR | 6 | 104 | 124 | 0.55 | This study |
| Zeuxine strateumatica | RAPD | 10 | 50 | 71 | 0.92 | [34] |
| Eulophia sinensis | RAPD | 7 | 38 | 97 | 0.65 | [34] |
| Zeuxine gracilis | RAPD | 6 | 74 | 77 | 0.54 | [34] |
| Goodyera procera | RAPD | 14 | 343 | 101 | 0.39 | [35] |
| Platanthera leucophaea | RAPD | 10 | 192 | 64 | 0.26 | [36] |
| Paphiopedilum micranthum | RAPD | 4 | 161 | 131 | 0.20 | [37] |
| Changnienia amoena | RAPD | 11 | 216 | 119 | 0.43 | [38] |
| Cattleya labiata | RAPD and ISSR | 7 | 117 | 272 | 0.13 | [26] |
| Vanda coerulea | RAPD and ISSR | 7 | 32 | 226 | 0.04 | [39] |
| Liparis loeselii | AFLP | 12 | 155 | 108 | 0.38 | [5] |
| Dendrobium officinale | AFLP | 12 | 71 | 195 | 0.27 | [18] |
| Liparis japonica | AFLP | 8 | 185 | 406 | 0.43 | [19] |
| Spiranthes romanzoffiana | AFLP | 17 | 205 | 138 | 0.89 | [40] |
| Phragmipedium longifolium | AFLP | 6 | 160 | 365 | 0.20 | [41] |
| Himantoglossum hircinum | AFLP | 20 | 211 | 215 | 0.15 | [42] |
| Orchis mascula | AFLP | 15 | 293 | 196 | 0.08 | [43] |
| Orchis purpurea | AFLP | 9 | 244 | 70 | 0.09 | [44] |
| Dactylorhiza incarnata | AFLP | 12 | 250 | 91 | 0.35 | [45] |
| Cypripedium japonicum | AFLP | 6 | 180 | 377 | 0.52 | [46] |
| Dendrobium loddigesii | SRAP | 7 | 92 | 231 | 0.30 | [20] |
| Population Code | Locality | Geographical Coordinate | Altitude (m) | Sample Size | Voucher Number |
|---|---|---|---|---|---|
| LA | Linan county, Zhejiang | 30°21′N,119°25′E | 1021 | 20 | J0611 |
| RJ | Rongjiang county, Guizhou | 26°23′N,108°12′E | 516 | 20 | Q0516 |
| SZ | Sangzhi county, Hunan | 29°40′N,110°2′E | 363 | 20 | Q0605 |
| TR | Tongren county, Guizhou | 27°56′N,180°36′E | 818 | 15 | Q0521 |
| WN | Wuning county, Jiangxi | 29°6′N,115°17′E | 582 | 9 | Q0418 |
| WYS | Wuyishan county, Fujian | 27°44′N,117°41′E | 705 | 20 | Q0507 |
| Primer Name | Sequence (5′→3′) | Annealing Temperature (°C) |
|---|---|---|
| UBC813 | (CT)8T | 50 |
| UBC818 | (CA)8G | 52 |
| UBC824 | (TC)8G | 51 |
| UBC828 | (TG)8A | 51 |
| UBC834 | (AG)8YT | 50 |
| UBC843 | (CT)8RA | 50 |
| UBC845 | (CT)8RG | 52 |
| UBC859 | (CG)8RC | 52 |
| UBC868 | (GAA)6 | 48 |
| UBC873 | (GACA)4 | 49 |
| UBC881 | (GGGTG)3 | 56 |
Acknowledgments
Conflicts of Interest
References
- Swarts, N.D.; Dixon, K.W. Terrestrial orchid conservation in the age of extinction. Ann. Bot 2009, 104, 543–556. [Google Scholar]
- Kim, S.H.; Lee, J.S.; Lee, G.J.; Kim, J.S.; Ha, B.K.; Kim, D.S.; Kim, J.B.; Kang, S.Y. Analyses of genetic diversity and relationships in four Calanthe taxa native to Korea using AFLP markers. Hort. Environ. Biotechnol 2013, 54, 148–155. [Google Scholar]
- Chen, X.Q.; Ji, Z.H.; Lang, K.Y.; Zhu, G.H. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1999; Volume 18, pp. 279–281. [Google Scholar]
- Subject Database of China Plant. Available online: http://www.plant.csdb.cn/protectlist (accessed on 18 August 2013).
- Pillon, Y.; Qamaruz-Zaman, F.; Fay, M.F.; Hendoux, F.; Piquot, Y. Genetic diversity and ecological differentiation in the endangered fen orchid (Liparis loeselii). Conserv. Genet 2007, 8, 177–184. [Google Scholar]
- Izawa, T.; Kawahara, T.; Takahashi, H. Genetic diversity of an endangered plant, Cypripedium macranthos var. rebunense (Orchidaceae): Background genetic research for future conservation. Conserv. Genet 2007, 8, 1369–1376. [Google Scholar]
- George, S.; Sharma, J.; Yadon, V.L. Genetic diversity of the endangered and narrow endemic Piperia yadonii (Orchidaceae) assessed with ISSR polymorphisms. Am. J. Bot 2009, 96, 2022–2030. [Google Scholar]
- Rubio-Moraga, A.; Candel-Perez, D.; Lucas-Borja, M.E.; Tiscar, P.A.; Viñegla, B.; Linares, J.C.; Gómez-Gómez, L.; Ahrazem, O. Genetic diversity of Pinus nigra Arn. populations in Southern Spain and Northern Morocco revealed by Inter-Simple Sequence Repeat profiles. Int. J. Mol. Sci 2012, 13, 5645–5658. [Google Scholar]
- Farsani, T.M.; Etemadi, N.; Sayed-Tabatabaei, B.E.; Talebi, M. Assessment of genetic diversity of Bermudagrass (Cynodon dactylon) using ISSR markers. Int. J. Mol. Sci 2012, 13, 383–392. [Google Scholar]
- Golkar, P.; Arzani, A.; Rezaei, A.M. Genetic variation in Safflower (Carthamus tinctorious L.) for seed quality-related traits and Inter-Simple Sequence Repeat (ISSR) markers. Int. J. Mol. Sci 2011, 12, 2664–2677. [Google Scholar]
- Yao, X.H.; Gao, L.; Yang, B. Genetic diversity of wild Cymbidium goeringii (Orchidaceae) populations from Hubei based on inter-simple sequence repeats analysis. Front. Biol. China 2007, 2, 419–424. [Google Scholar]
- Smith, J.L.; Hunter, K.L.; Hunter, R.B. Genetic variation in the terrestrial orchid Tipularia discolor. Southeast. Nat 2002, 1, 17–26. [Google Scholar]
- Wallace, L.E. A comparison of genetic variation and structure in the allopolyploid Platanthera huronensis and its diploid progenitors, Platanthera aquilonis and Platanthera dilatata. Can. J. Bot 2004, 82, 244–252. [Google Scholar]
- Wu, H.F.; Li, Z.Z.; Huang, H.W. Genetic differentiation among natural populations of Gastrodia elata (Orchidaceae) in Hubei and germplasm assessment of the cultivated populations. Biodivers. Sci 2006, 14, 315–326. [Google Scholar]
- Ma, J.M.; Yin, S.H. Genetic diversity of Dendrobium fimbriatum (Orchidaceae), an endangered species, detected by inter-simple sequence repeat (ISSR). Acta Bot. Yunn 2009, 31, 35–41. [Google Scholar]
- Wei, L.; Wu, X.J. Genetic variation and population differentiation in a medical herb Houttuynia cordata in China revealed by Inter-Simple Sequence Repeats (ISSRs). Int. J. Mol. Sci 2012, 13, 8159–8170. [Google Scholar]
- Chung, M.Y.; López-Pujol, J.; Maki, M.; Moon, M.O.; Hyun, J.O.; Chung, M.G. Genetic variation and structure within 3 endangered Calanthe Species (Orchidaceae) from Korea: Inference of population-establishment history and implications for conservation. J. Hered 2013, 104, 248–262. [Google Scholar]
- Li, X.X.; Ding, X.Y.; Chu, B.H.; Zhou, Q.; Ding, G.; Gu, S. Genetic diversity analysis and conservation of the endangered Chinese endemic herb Dendrobium officinale Kimura et Migo (Orchidaceae) based on AFLP. Genetica 2008, 133, 159–166. [Google Scholar]
- Chen, X.H.; Guan, J.J.; Ding, R.; Zhang, Q.; Ling, X.Z.; Qu, B.; Zhang, L.J. Conservation genetics of the endangered terrestrial orchid Liparis japonica in Northeast China based on AFLP markers. Plant Syst. Evol 2013, 299, 691–698. [Google Scholar]
- Cai, X.Y.; Feng, Z.Y.; Zhang, X.X.; Xu, W.; Hou, B.W.; Ding, X.Y. Genetic diversity and population structure of an endangered orchid (Dendrobium loddigesii Rolfe) from China revealed by SRAP markers. Sci. Hortic 2011, 129, 877–881. [Google Scholar]
- Yu, H.H.; Yang, Z.L.; Sun, B.; Liu, R.N. Genetic diversity and relationship of endangered plant Magnolia officinalis (Magnoliaceae) assessed with ISSR polymorphisms. Biochem. Syst. Ecol 2011, 39, 71–78. [Google Scholar]
- Gonzalez-astorga, J.; Castillo-campos, G. Genetic variability of the narrow endemic tree Antirhea aromatica Castillo-Campos & Lorence, (Rubiaceae,Guettardeae) in a tropical forest of Mexico. Ann. Bot 2004, 93, 521–528. [Google Scholar]
- Li, J.M.; Jin, Z.X. Genetic variation and differentiation in Torreya jackii Chun, an endangered plant endemic to China. Plant Sci 2007, 172, 1048–1053. [Google Scholar]
- Hamrick, J.L.; Godt, M.J.W. Allozyme Diversity in Plant Species. In Plant Population Genetics, Breeding, and Genetic Resources; Brown, A.H.D., Clegg, M.T., Kahler, A.L., Weir, B.S., Eds.; Sinauer Associates: Sunderland, MA, USA, 1990; pp. 43–63. [Google Scholar]
- Shen, L.; Chen, X.Y.; Li, Y.Y. Glacial refugia and postglacial recolonization patterns of organisms. Acta Ecol. Sin 2002, 22, 1983–1990. [Google Scholar]
- Pinheiro, L.R.; Rabbani, A.R.C.; da Silva, A.V.C.; da Silva Lédo, A.; Pereira, K.L.G.; Diniz, L.E.C. Genetic diversity and population structure in the Brazilian Cattleya labiata (Orchidaceae) using RAPD and ISSR markers. Plant Syst. Evol 2012, 298, 1815–1825. [Google Scholar]
- Zhao, X.F.; Ma, Y.P.; Sun, W.B.; Wen, X.Y.; Milne, R. High genetic diversity and low differentiation of Michelia coriacea (Magnoliaceae), a critically endangered endemic in Southeast Yunnan, China. Int. J. Mol. Sci 2012, 13, 4396–4411. [Google Scholar]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol 2004, 13, 1143–1155. [Google Scholar]
- Barrett, S.C.H.; Kohn, J.R. Genetic and Evolutionary Consequences of Small Population Size in Plants: Implications for Conservation. In In Genetics and Conservation of Rare Plants; Falk, D.A., Holsinger, K.E., Eds.; Oxford University Press: New York, NY, USA, 1991; pp. 3–30. [Google Scholar]
- Brzosko, E.; Wróblewska, A.; Tałałaj, I.; Wasilewska, E. Genetic diversity of Cypripedium calceolus in Poland. Plant Syst. Evol 2011, 295, 83–96. [Google Scholar]
- Liu, J.F.; Shi, S.Q.; Chang, E.M.; Yang, W.J.; Jiang, Z.P. Genetic diversity of the critically endangered Thuja sutchuenensis revealed by ISSR markers and the implications for conservation. Int. J. Mol. Sci 2013, 14, 14860–14871. [Google Scholar]
- Eduardo, L.B.; Joaa, O.S.; George, J.S. Self-incompatibility, inbreeding depression and crossing potential in five Brazilian Pleurothallis (Orchidaceae) Species. Ann. Bot 2001, 88, 89–99. [Google Scholar]
- Lian, J.J.; Qian, X.; Wang, C.X.; Liu, F.; Tian, M; Tang, T; Wang, F.T. Ovule development and seed formation of Calanthe tsoongiana. Acta Bot. Boreal.-Occident. Sin 2013, 33, 494–500. [Google Scholar]
- Sun, M.; Wong, K.C. Genetic structure of three orchid species with contrasting breeding systems using RAPD and allozyme markers. Am. J. Bot 2001, 88, 2180–2188. [Google Scholar]
- Wong, K.C.; Sun, M. Reproductive biology and conservation genetics of Goodyera procera (Orchidaceae). Am. J. Bot 1999, 86, 1406–1413. [Google Scholar]
- Wallace, L.E. Examining the effects of fragmentation on genetic variation in Platanthera leucophaea (Orchidaceae): Inferences from allozyme and random amplified polymorphic DNA markers. Plant Spec. Biol 2002, 17, 37–49. [Google Scholar]
- Li, A.; Luo, Y.B.; Ge, S. A preliminary study on conservation genetics of an endangered orchid (Paphiopedilum micranthum) from Southwestern China. Biochem. Genet 2002, 40, 195–201. [Google Scholar]
- Li, A.; Ge, S. Genetic variation and conservation of Changnienia amoena, an endangered orchid endemic to China. Plant Syst. Evol 2006, 258, 251–260. [Google Scholar]
- Manners, V.; Kumaria, S.; Tandon, P. SPAR methods revealed high genetic diversity within populations and high gene flow of Vanda coerulea Griff ex Lindl (Blue Vanda), an endangered orchid species. Gene 2013, 519, 91–97. [Google Scholar]
- Forrest, A.D.; Hollingsworth, M.L.; Hollingsworth, P.M.; Sydes, C.; Bateman, R.M. Population genetic structure in European populations of Spiranthes romanzoffi ana set in the context of other genetic studies on orchids. Heredity 2004, 92, 218–227. [Google Scholar]
- Muñoz, M.; Warner, J.; Albertazzi, F.J. Genetic diversity analysis of the endangered slipper orchid Phragmipedium longifolium in Costa Rica. Plant Syst. Evol 2010, 290, 217–223. [Google Scholar]
- Pfeifer, M.; Schatz, B.; Picó, F.X.; Passalacqua, N.G.; Fay, M.F.; Carey, P.D.; Jeltsch, F. Phylogeography and genetic structure of the orchid Himantoglossum hircinum (L.) Spreng. across its European central–marginal gradient. J. Biogeogr 2009, 36, 2353–2365. [Google Scholar]
- Jacquemyn, H.; Brys, R.; Adriaens, D.; Honnay, O.; Roldán-Ruiz, I. Effects of population size and forest management on genetic diversity and structure of the tuberous orchid Orchis mascula. Conserv. Genet 2009, 10, 161–168. [Google Scholar]
- Jacquemyn, H.; Vandepitte, K.; Brys, R.; Honnay, O.; Roldán-Ruiz, I. Fitness variation and genetic diversity in small, remnant populations of the food deceptive orchid Orchis purpurea. Biol. Conserv 2007, 139, 203–210. [Google Scholar]
- Vandepitte, K.; Gristina, A.S.; de Hert, K.; Meekers, T.; Roldán-ruiz, I.; Honnay, O. Recolonization after habitat restoration leads to decreased genetic variation in populations of a terrestrial orchid. Mol. Ecol 2012, 21, 4206–4215. [Google Scholar]
- Qian, X.; Li, Q.J.; Lian, J.J.; Wang, C.X.; Tian, M. Genetic diversity of endangered wild Cypripedium japonicum populations: An AFLP analysis. Chinese J. Ecol 2013, 32, 1445–1450. [Google Scholar]
- Wright, S. The genetical structure of populations. Ann. Eugenic 1949, 15, 323–354. [Google Scholar]
- Fay, M.F.; Bone, R.; Cook, P.; Kahandawala, I.; Greensmith, J.; Harris, S.; Pedersen, H.; Ingrouille, M.J.; Lexer, C. Genetic diversity in Cypripedium calceolus (Orchidaceae) with a focus on north-western Europe, as revealed by plastid DNA length polymorphisms. Ann. Bot 2009, 104, 517–525. [Google Scholar]
- Dodd, S.C.; Helenurm, K. Genetic diversity in Delphinium variegatum (Ranunculaceae): A comparison of two insular endemic subspecies and their widespread mainland relative. Am. J. Bot 2002, 89, 613–622. [Google Scholar]
- Godt, M.J.W.; Hamrick, J.L. Allozyme variation in two Great Smoky Mountain endemics: Cacalia regalia and Glyceria nubigena. J. Hered 1995, 86, 194–198. [Google Scholar]
- POPGENE, version 1.32; Software for population genetic analysis; University of Alberta: Edmonton, AB, Canada, 1999.
- NTSYS-pc, Numerical Taxonomy and Multivariate Analysis System, version 2.2; Exeter Software: Setauket, NY, USA, 2005.
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar]
- Tools for Population Genetic Analyses (TFPGA), version 1.3. In A Windows program for the analysis of allozyme and molecular population genetic data; Utah State University: Logan, UT, USA, 1997.
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure from multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour 2012, 4, 359–361. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol 2005, 14, 2611–2620. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Qian, X.; Wang, C.-x.; Tian, M. Genetic Diversity and Population Differentiation of Calanthe tsoongiana, a Rare and Endemic Orchid in China. Int. J. Mol. Sci. 2013, 14, 20399-20413. https://doi.org/10.3390/ijms141020399
Qian X, Wang C-x, Tian M. Genetic Diversity and Population Differentiation of Calanthe tsoongiana, a Rare and Endemic Orchid in China. International Journal of Molecular Sciences. 2013; 14(10):20399-20413. https://doi.org/10.3390/ijms141020399
Chicago/Turabian StyleQian, Xin, Cai-xia Wang, and Min Tian. 2013. "Genetic Diversity and Population Differentiation of Calanthe tsoongiana, a Rare and Endemic Orchid in China" International Journal of Molecular Sciences 14, no. 10: 20399-20413. https://doi.org/10.3390/ijms141020399