MiR-205 Is Progressively Down-Regulated in Lymph Node Metastasis but Fails as a Prognostic Biomarker in High-Risk Prostate Cancer
Abstract
:1. Introduction
2. Results
2.1. MiR-205 Is Under-Expressed and Its Reconstitution Leads to Proliferation—Inhibition in PCa Cells
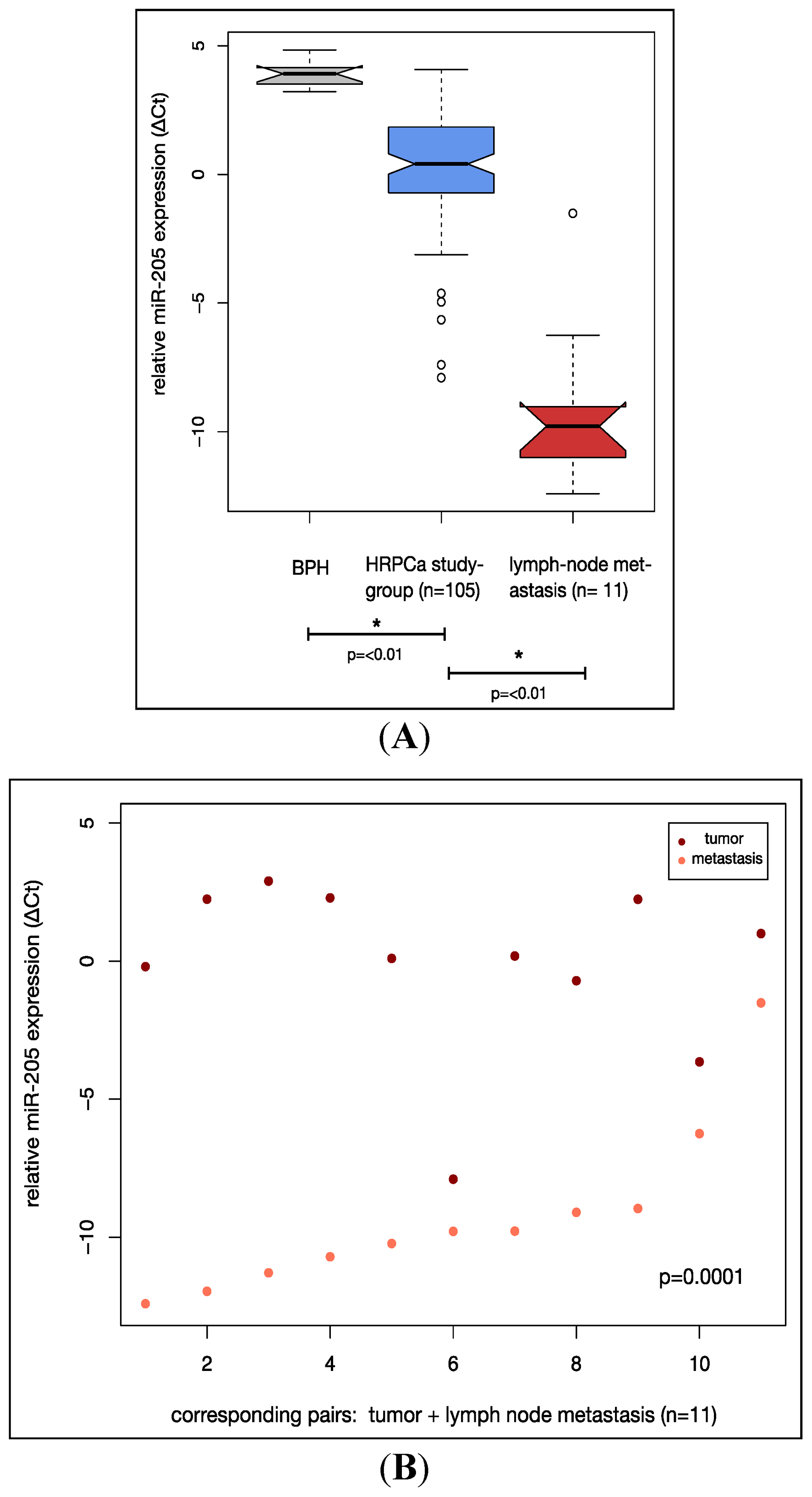
2.2. MiR-205 Is Under-Expressed in a HRPCa and Shows Increasing Down-Regulation in Lymph Node Metastases
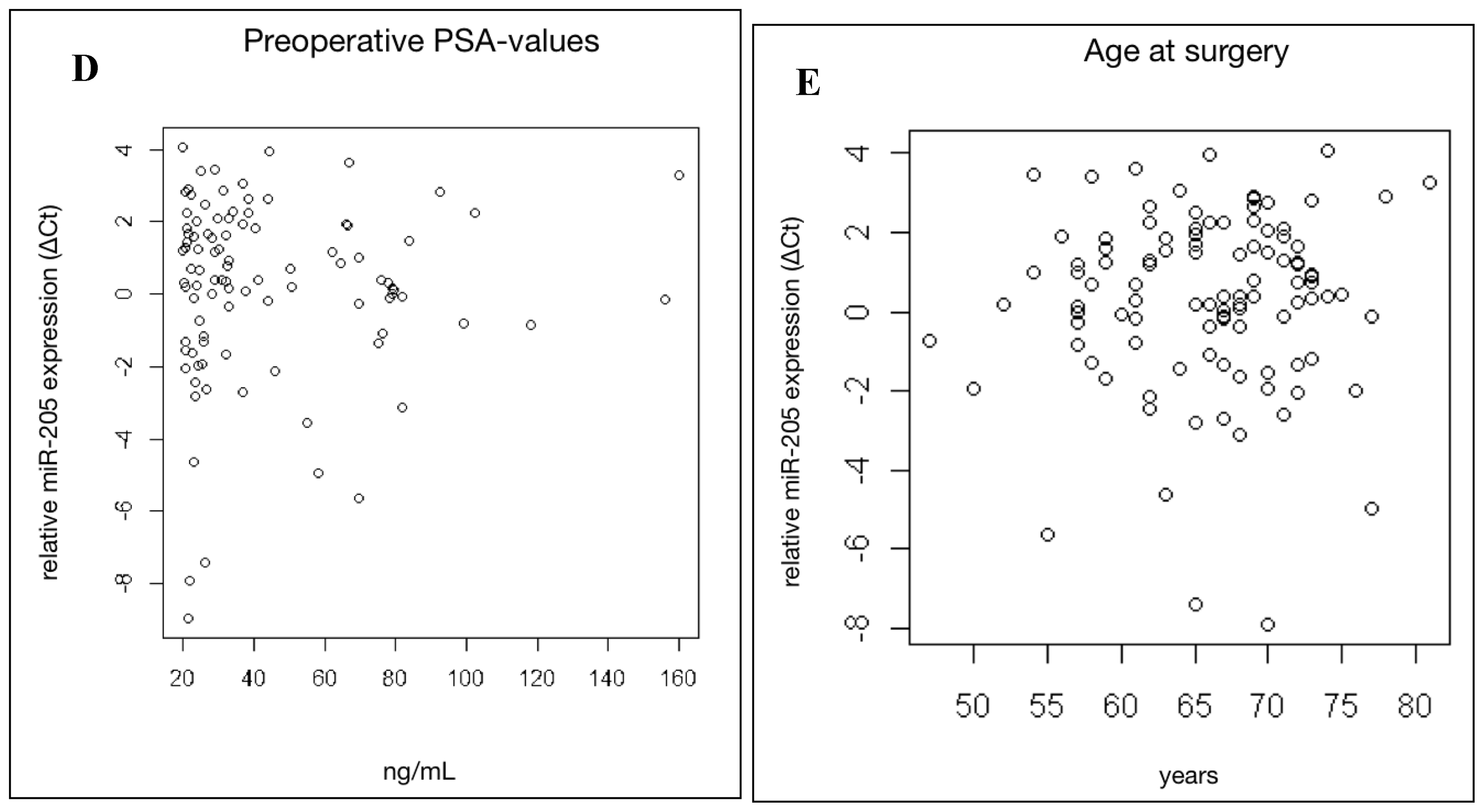
2.3. MiR-205 Expression Is Not Significantly Linked to Clinical Prognostic Parameters in HRPCa
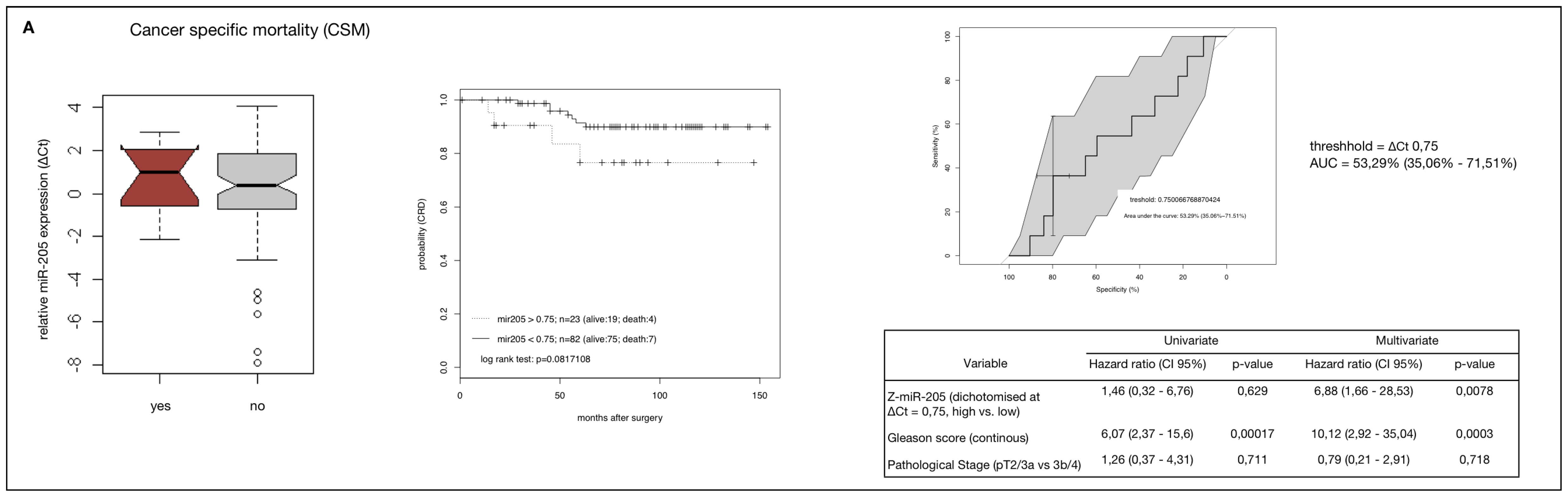
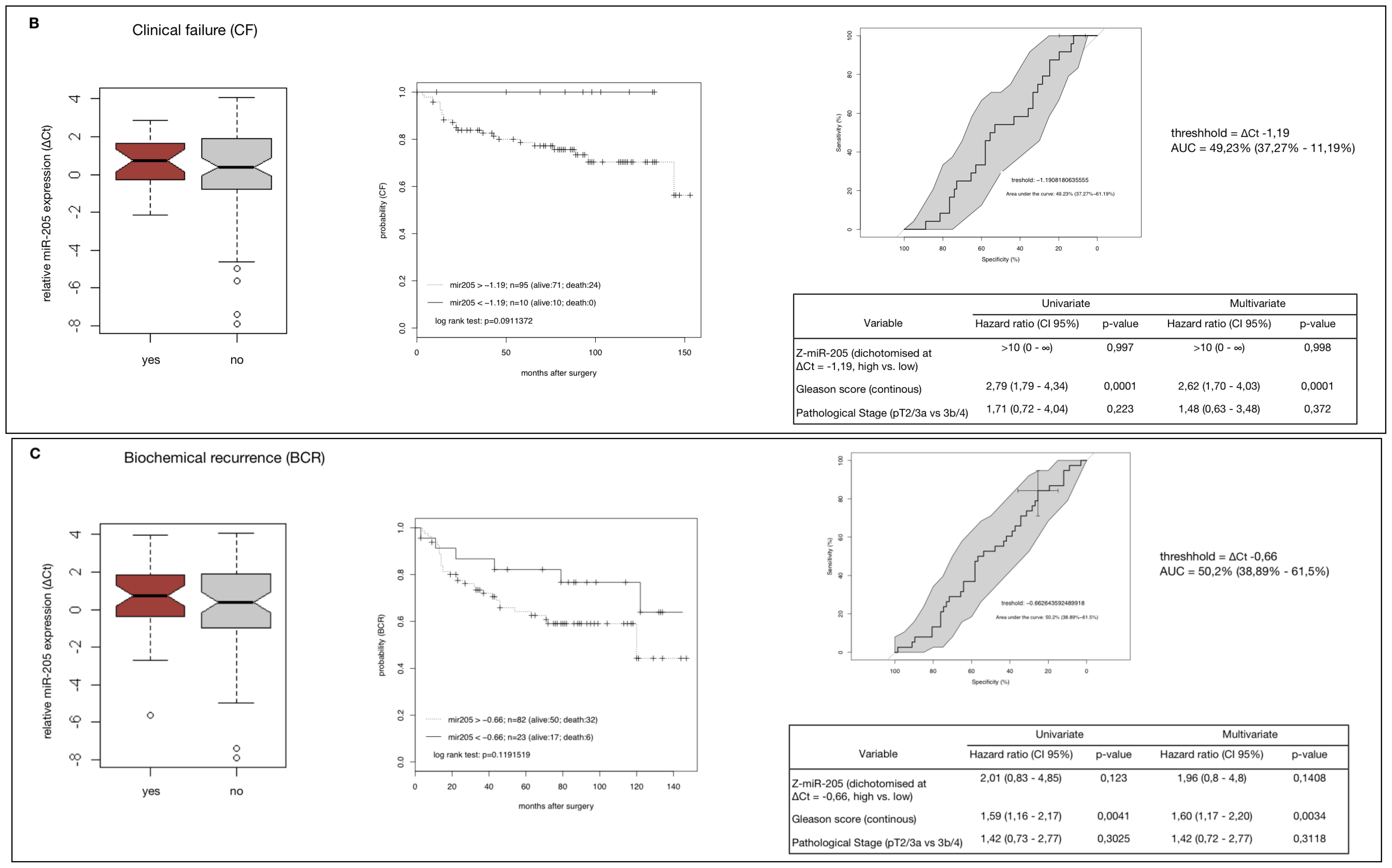
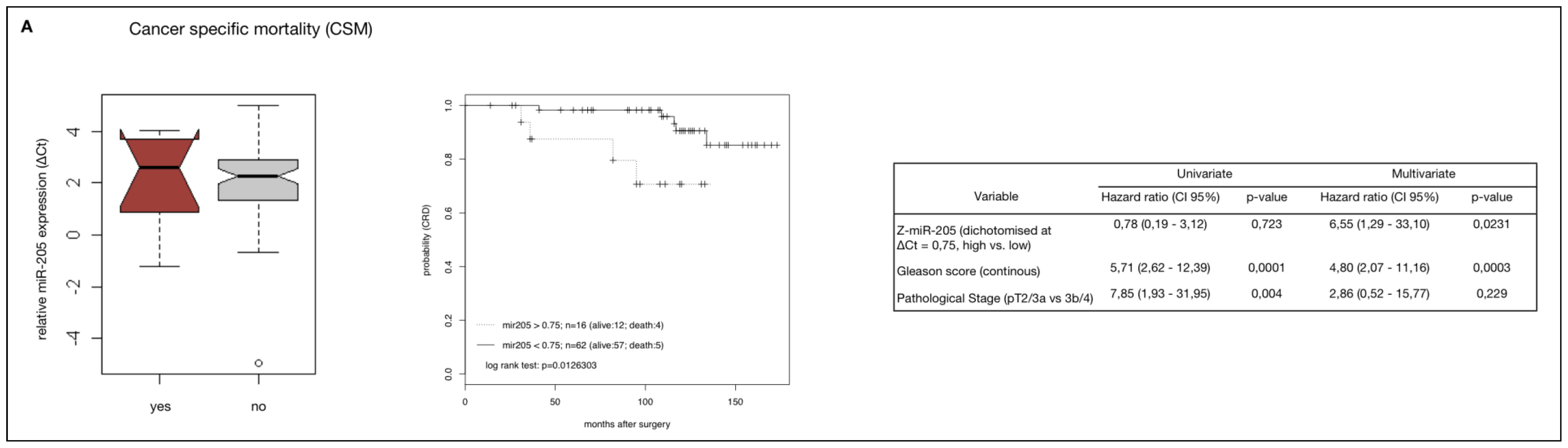
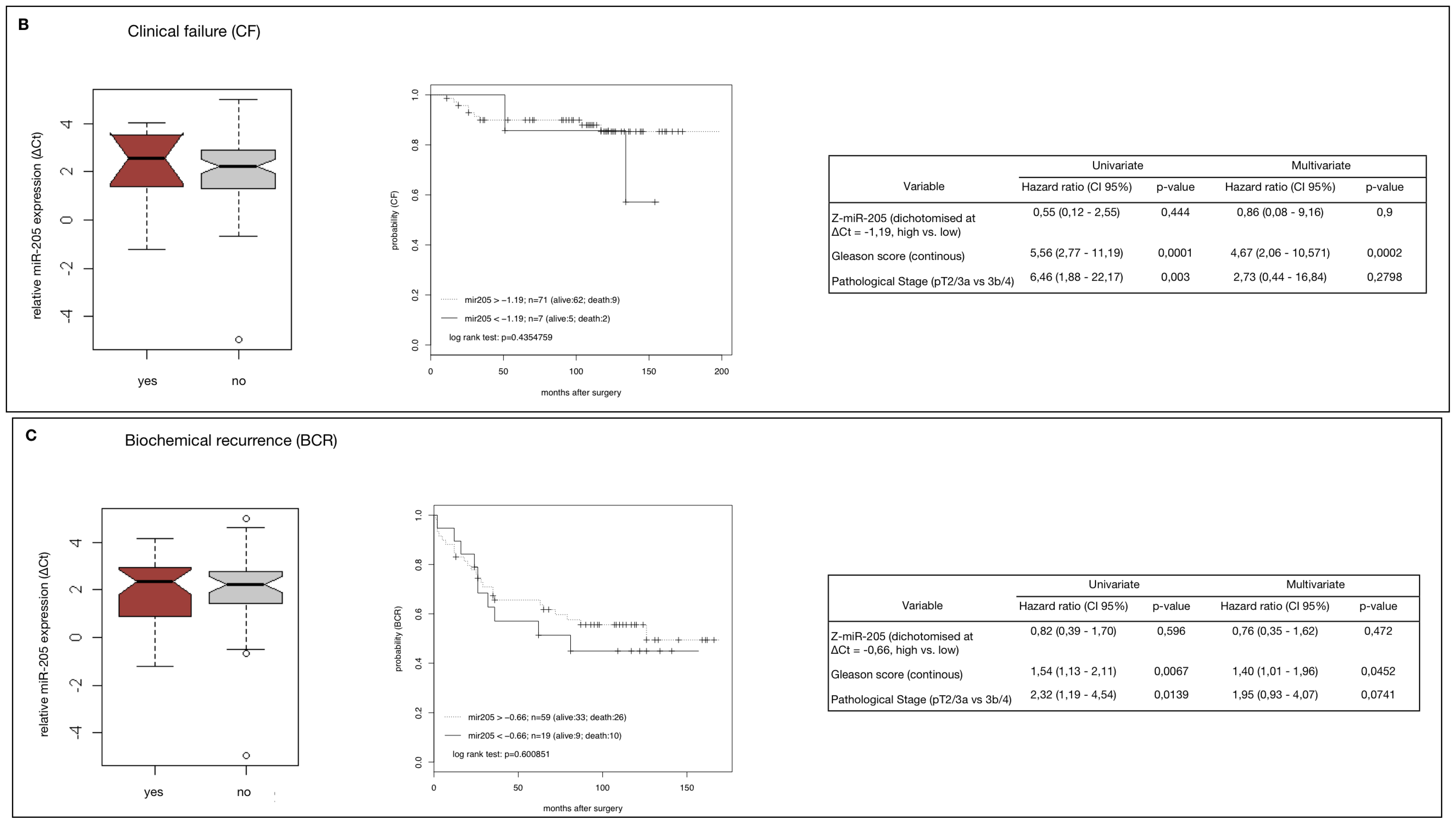
2.4. MiR-205 as a Prognostic Factor in HRPCa
2.5. Down-Regulation of MiR-205 Fails as a Prognostic Factor in HRPCa
3. Discussion
4. Experimental Section
4.1. Cell Lines
4.2. Cell Transfection
4.3. Cell Proliferation Assay
4.4. In Vitro Invasion Assay
4.5. Patients and Samples
4.6. qRT-PCR
4.7. Statistical and Bioinformatical Analysis
5. Conclusions
Supplementary Information
ijms-14-21414-s001.pdf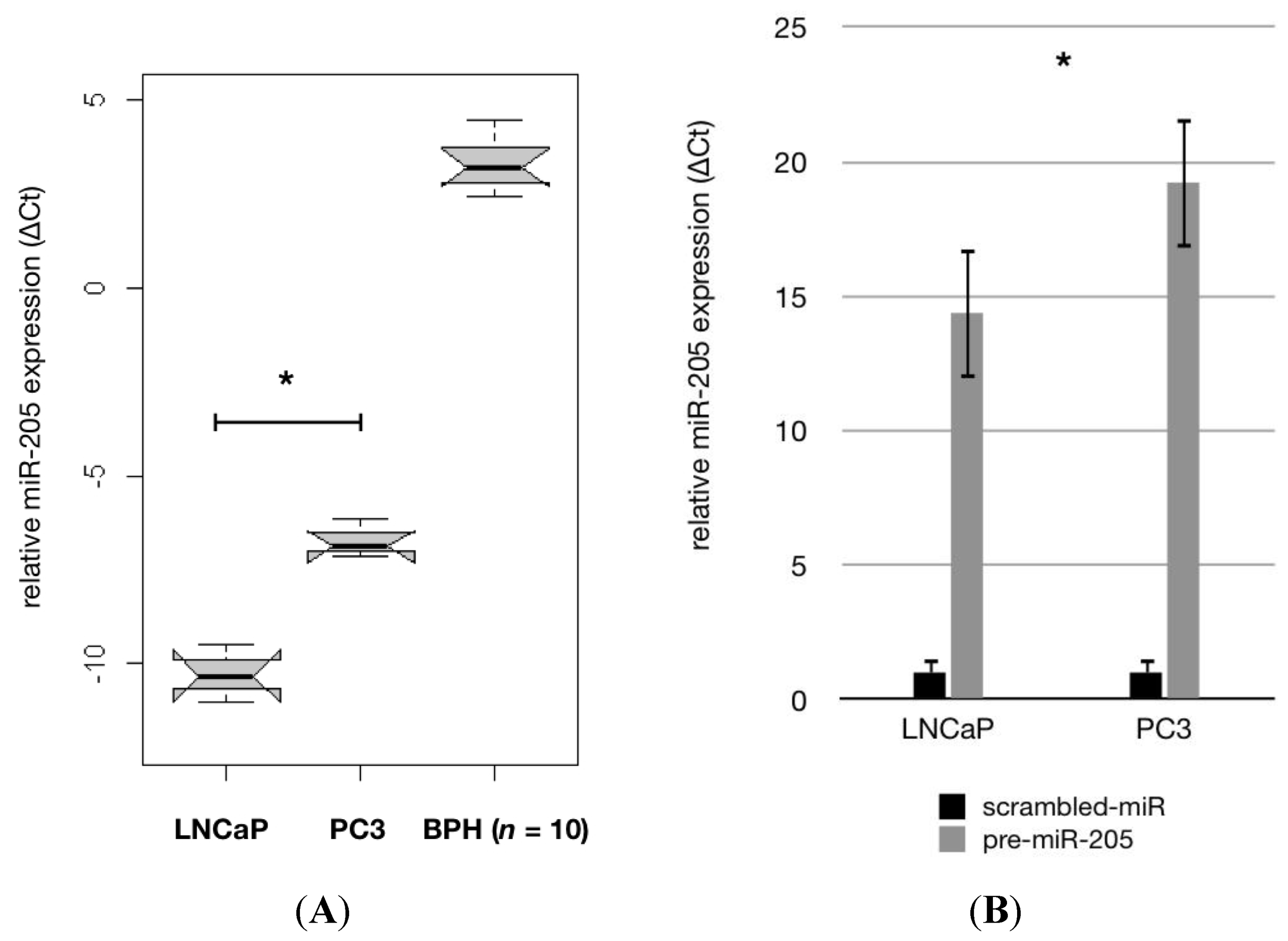


| Study cohort | Validation cohort | |
|---|---|---|
| Number of patients | 105 | 78 |
| Median ΔCt miR-205 expression (range) | 0.33 (−7.9–4.1) | 1.96 (−4.95–5.0) |
| Median follow-up, months (range) | 78.3 (1–154) | 117 (14–173) |
| Median Age, years (range) | 65.9 (47–81) | 63 (41–75) |
| Median PSA, ng/μL (range) | 32 (20–160) | 14 (2.7–95.3) |
| Gleason Score (GS): | ||
| GS ≤ 6 | 2 (1.9%) | 24 (30.4%) |
| GS =7 | 31 (29.5%) | 37 (46.8%) |
| GS 8–10 | 72 (68.6%) | 18 (22.8%) |
| Pathological Stage (pT): | ||
| pT 2 | 17 (17.2%) | 24 (34.2%) |
| pT3a | 28 (26.7%) | 35 (44.3%) |
| pT3b | 44 (41.9%) | 16 (24.1%) |
| pT4 | 16 (15.2%) | 4 (5.1%) |
| Nodal Status (LN): | ||
| LN neg | 64 (61.0%) | 70 (89.7%) |
| LN pos | 41 (39.0%) | 8 (10.3%) |
| Relapse Events: | ||
| Biochemical recurrence | 38 (36.2%) | 36 (49.4%) |
| Clinical failure | 24 (22.9%) | 11 (13.9%) |
| Cancer-specific mortality | 11 (10.5%) | 9 (11.4%) |
| Any cause death | 21 (20.0%) | 16 (24.1%) |
Acknowledgments
Conflicts of Interest
References
- Grimm, P.; Billiet, I.; Bostwick, D.; Dicker, A.P.; Frank, S.; Immerzeel, J.; Keyes, M.; Kupelian, P.; Lee, W.R.; Machtens, S.; et al. Comparative analysis of prostate-specific antigen free survival outcomes for patients with low, intermediate and high risk prostate cancer treatment by radical therapy. Results from the prostate cancer results study group. BJU Int 109 Suppl 2012, 1, 22–29. [Google Scholar]
- Bastian, P.J.; Boorjian, S.A.; Bossi, A.; Briganti, A.; Heidenreich, A.; Freedland, S.J.; Montorsi, F.; Roach, M., 3rd; Schroder, F.; van Poppel, H.; et al. High-risk prostate cancer: From definition to contemporary management. Eur. Urol 2012, 61, 1096–1106. [Google Scholar]
- Harisinghani, M.G.; Weissleder, R. Sensitive, noninvasive detection of lymph node metastases. PLoS Med 2004, 1, e66. [Google Scholar]
- Choudhury, A.D.; Eeles, R.; Freedland, S.J.; Isaacs, W.B.; Pomerantz, M.M.; Schalken, J.A.; Tammela, T.L.; Visakorpi, T. The role of genetic markers in the management of prostate cancer. Eur. Urol 2012, 62, 577–587. [Google Scholar]
- Kim, T.; Reitmair, A. Non-coding RNAs: Functional aspects and diagnostic utility in oncology. Int. J. Mol. Sci 2013, 14, 4934–4968. [Google Scholar]
- Du, T.; Zamore, P.D. Microprimer: The biogenesis and function of microRNA. Development 2005, 132, 4645–4652. [Google Scholar]
- Meltzer, P.S. Cancer genomics: Small RNAs with big impacts. Nature 2005, 435, 745–746. [Google Scholar]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar]
- Huang, Q.; Gumireddy, K.; Schrier, M.; le Sage, C.; Nagel, R.; Nair, S.; Egan, D.A.; Li, A.; Huang, G.; Klein-Szanto, A.J.; et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol 2008, 10, 202–210. [Google Scholar]
- Spahn, M.; Kneitz, S.; Scholz, C.J.; Stenger, N.; Rudiger, T.; Strobel, P.; Riedmiller, H.; Kneitz, B. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int. J. Cancer 2009, 127, 394–403. [Google Scholar]
- Ambs, S.; Prueitt, R.L.; Yi, M.; Hudson, R.S.; Howe, T.M.; Petrocca, F.; Wallace, T.A.; Liu, C.G.; Volinia, S.; Calin, G.A.; et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 2008, 68, 6162–6170. [Google Scholar]
- Porkka, K.P.; Pfeiffer, M.J.; Waltering, K.K.; Vessella, R.L.; Tammela, T.L.; Visakorpi, T. MicroRNA expression profiling in prostate cancer. Cancer Res 2007, 67, 6130–6135. [Google Scholar]
- Ozen, M.; Creighton, C.J.; Ozdemir, M.; Ittmann, M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008, 27, 1788–1793. [Google Scholar]
- Schaefer, A.; Jung, M.; Mollenkopf, H.J.; Wagner, I.; Stephan, C.; Jentzmik, F.; Miller, K.; Lein, M.; Kristiansen, G.; Jung, K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int. J. Cancer 2010, 126, 1166–1176. [Google Scholar]
- Qin, A.Y.; Zhang, X.W.; Liu, L.; Yu, J.P.; Li, H.; Wang, S.Z.; Ren, X.B.; Cao, S. MiR-205 in cancer: An angel or a devil? Eur. J. Cell Biol 2013, 92, 54–60. [Google Scholar]
- Greene, S.B.; Herschkowitz, J.I.; Rosen, J.M. The ups and downs of miR-205: Identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol 2010, 7, 300–304. [Google Scholar]
- Hagman, Z.; Haflidadottir, B.S.; Ceder, J.A.; Larne, O.; Bjartell, A.; Lilja, H.; Edsjo, A.; Ceder, Y. MiR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br. J. Cancer 2013, 108, 1668–1676. [Google Scholar]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar]
- Boll, K.; Reiche, K.; Kasack, K.; Morbt, N.; Kretzschmar, A.K.; Tomm, J.M.; Verhaegh, G.; Schalken, J.; von Bergen, M.; Horn, F.; et al. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 2012, 32, 277–285. [Google Scholar]
- Iorio, M.V.; Casalini, P.; Piovan, C.; Di Leva, G.; Merlo, A.; Triulzi, T.; Menard, S.; Croce, C.M.; Tagliabue, E. MicroRNA-205 regulates her3 in human breast cancer. Cancer Res 2009, 69, 2195–2200. [Google Scholar]
- Wu, H.; Zhu, S.; Mo, Y.Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 2009, 19, 439–448. [Google Scholar]
- Dijckmeester, W.A.; Wijnhoven, B.P.; Watson, D.I.; Leong, M.P.; Michael, M.Z.; Mayne, G.C.; Bright, T.; Astill, D.; Hussey, D.J. MicroRNA-143 and -205 expression in neosquamous esophageal epithelium following argon plasma ablation of barrett’s esophagus. J. Gastrointest. Surg 2009, 13, 846–853. [Google Scholar]
- Gandellini, P.; Folini, M.; Longoni, N.; Pennati, M.; Binda, M.; Colecchia, M.; Salvioni, R.; Supino, R.; Moretti, R.; Limonta, P.; et al. MiR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase cepsilon. Cancer Res 2009, 69, 2287–2295. [Google Scholar]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting zeb1 and sip1. Nat. Cell Biol 2008, 10, 593–601. [Google Scholar]
- Puhr, M.; Hoefer, J.; Schafer, G.; Erb, H.H.; Oh, S.J.; Klocker, H.; Heidegger, I.; Neuwirt, H.; Culig, Z. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am. J. Pathol 2012, 181, 2188–2201. [Google Scholar]
- Tucci, P.; Agostini, M.; Grespi, F.; Markert, E.K.; Terrinoni, A.; Vousden, K.H.; Muller, P.A.; Dotsch, V.; Kehrloesser, S.; Sayan, B.S.; et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 15312–15317. [Google Scholar]
- Hulf, T.; Sibbritt, T.; Wiklund, E.D.; Patterson, K.; Song, J.Z.; Stirzaker, C.; Qu, W.; Nair, S.; Horvath, L.G.; Armstrong, N.J.; et al. Epigenetic-induced repression of microRNA-205 is associated with med1 activation and a poorer prognosis in localized prostate cancer. Oncogene 2013, 32, 2891–2899. [Google Scholar]
- Markou, A.; Tsaroucha, E.G.; Kaklamanis, L.; Fotinou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin. Chem 2008, 54, 1696–1704. [Google Scholar]
- Fletcher, A.M.; Heaford, A.C.; Trask, D.K. Detection of metastatic head and neck squamous cell carcinoma using the relative expression of tissue-specific miR-205. Transl. Oncol 2008, 1, 202–208. [Google Scholar]
- Vlietstra, R.J.; van Alewijk, D.C.; Hermans, K.G.; van Steenbrugge, G.J.; Trapman, J. Frequent inactivation of PTEN in prostate cancer cell lines and xenografts. Cancer Res 1998, 58, 2720–2723. [Google Scholar]
- Yu, J.; Peng, H.; Ruan, Q.; Fatima, A.; Getsios, S.; Lavker, R.M. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase ship2. FASEB J 2010, 24, 3950–3959. [Google Scholar]
- Schubert, M.; Spahn, M.; Kneitz, S.; Scholz, C.J.; Joniau, S.; Stroebel, P.; Kneitz, B. Distinct microRNA expression profile in prostate cancer patients with early clinical failure and the impact of let-7 as prognostic marker in high-risk prostate cancer. PLoS One 2013, 8, e65064. [Google Scholar]
- Ciafre, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun 2005, 334, 1351–1358. [Google Scholar]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar]
- Grunewald, T.G.; Kammerer, U.; Winkler, C.; Schindler, D.; Sickmann, A.; Honig, A.; Butt, E. Overexpression of lasp-1 mediates migration and proliferation of human ovarian cancer cells and influences zyxin localisation. Br. J. Cancer 2007, 96, 296–305. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kalogirou, C.; Spahn, M.; Krebs, M.; Joniau, S.; Lerut, E.; Burger, M.; Scholz, C.-J.; Kneitz, S.; Riedmiller, H.; Kneitz, B. MiR-205 Is Progressively Down-Regulated in Lymph Node Metastasis but Fails as a Prognostic Biomarker in High-Risk Prostate Cancer. Int. J. Mol. Sci. 2013, 14, 21414-21434. https://doi.org/10.3390/ijms141121414
Kalogirou C, Spahn M, Krebs M, Joniau S, Lerut E, Burger M, Scholz C-J, Kneitz S, Riedmiller H, Kneitz B. MiR-205 Is Progressively Down-Regulated in Lymph Node Metastasis but Fails as a Prognostic Biomarker in High-Risk Prostate Cancer. International Journal of Molecular Sciences. 2013; 14(11):21414-21434. https://doi.org/10.3390/ijms141121414
Chicago/Turabian StyleKalogirou, Charis, Martin Spahn, Markus Krebs, Steven Joniau, Evelyne Lerut, Maximilian Burger, Claus-Jürgen Scholz, Susanne Kneitz, Hubertus Riedmiller, and Burkhard Kneitz. 2013. "MiR-205 Is Progressively Down-Regulated in Lymph Node Metastasis but Fails as a Prognostic Biomarker in High-Risk Prostate Cancer" International Journal of Molecular Sciences 14, no. 11: 21414-21434. https://doi.org/10.3390/ijms141121414




