Characterization of a Maize Wip1 Promoter in Transgenic Plants
Abstract
:1. Introduction
2. Results
2.1. Isolation and Analysis of the Maize Wip1 Promoter
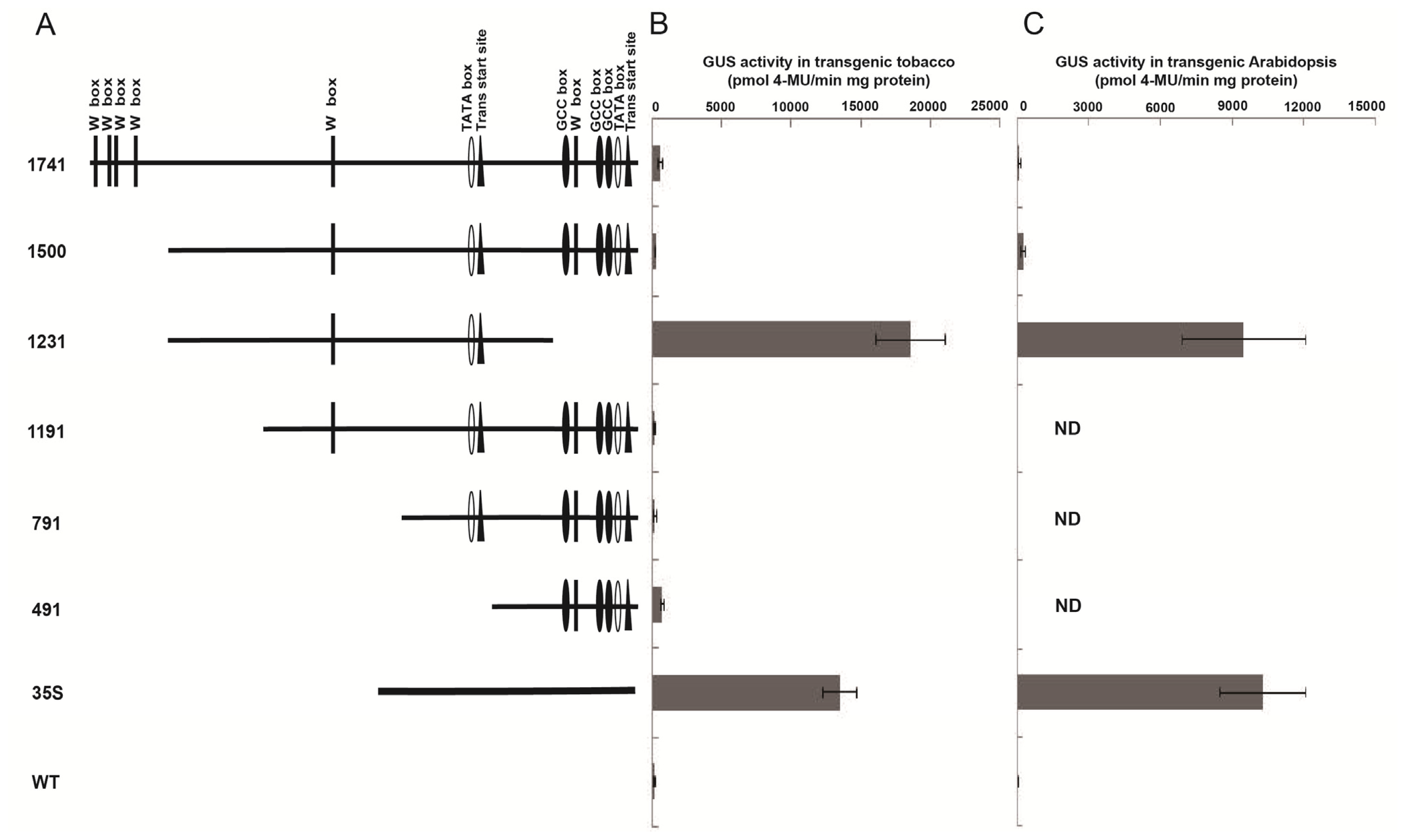
2.2. Truncated Wip1 Promoter Has Strong Activity in Transgenic Arabidopsis and Tobacco Leaves
2.3. Wip1 Promoters Are not Induced by Wounding and Have Different Activity in Different Organs in Transgenic Tobacco
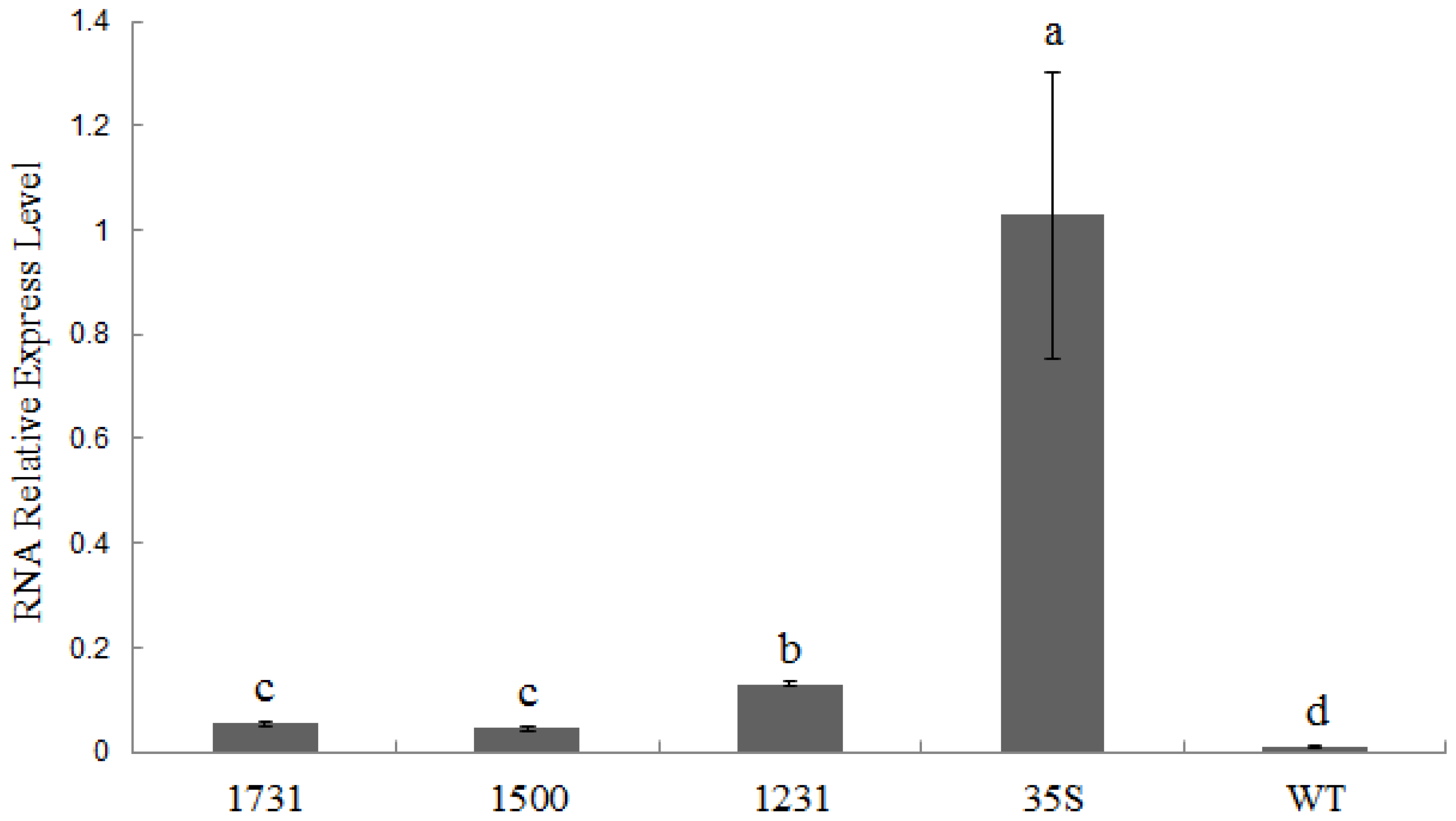
2.4. Wip1231 Promoter Drives Low GUS Transcriptional Level
2.5. Wip1 Promoters Have Multiple GUS Transcription Start Sites in Transgenic Tobacco
2.6. Wip1231 Promoters Were Influenced by the Adjacent 35S Promoter Sequence
2.7. Wip1 Promoter Is Wound Inducible in Transgenic Rice Plants
3. Discussion
4. Experimental Section
4.1. Isolation and Analysis of the Maize Wip1 Promoter
4.2. Plasmid Construction
4.3. Plant Transformation
4.4. Wounding of Transgenic Plant Leaves
4.5. Analysis of RNA Level by qRT-PCR
4.6. Analysis of the Transcription Start Sites by 5′ Rapid Amplification of cDNA Ends (5′-RACE)
4.7. GUS Activity Assay and Histochemical Staining
5. Conclusions
Supplementary Information
ijms-14-23872-s001.pdfAcknowledgments
Conflicts of Interest
References
- Fang, R.X.; Nagy, F.; Sivasubramaniam, S.; Chua, N.H. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell 1989, 1, 141–150. [Google Scholar]
- Christensen, A.H.; Sharrock, R.A.; Quail, P.H. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol 1992, 18, 675–689. [Google Scholar]
- Zhang, W.; McElroy, D.; Wu, R. Analysis of rice Act1 5′ region activity in transgenic rice plants. Plant Cell 1991, 3, 1155–1165. [Google Scholar]
- Cazzonelli, C.I.; McCallum, E.J.; Lee, R.; Botella, J.R. Characterization of a strong, constitutive mung bean (Vigna radiata L.) promoter with a complex mode of regulation in planta. Transgenic Res 2005, 14, 941–967. [Google Scholar]
- Stavolone, L.; Kononova, M.; Pauli, S.; Ragozzino, A.; de Haan, P.; Milligan, S.; Lawton, K.; Hohn, T. Cestrum yellow leaf curling virus (CmYLCV) promoter: A new strong constitutive promoter for heterologous gene expression in a wide variety of crops. Plant Mol. Biol 2003, 53, 663–673. [Google Scholar]
- Xie, Y.; Liu, Y.; Meng, M.; Chen, L.; Zhu, Z. Isolation and identification of a super strong plant promoter from cotton leaf curl Multan virus. Plant Mol. Biol 2003, 53, 1–14. [Google Scholar]
- Dynan, W.S. Modularity in promoters and enhancers. Cell 1989, 58, 1–4. [Google Scholar]
- Sunilkumar, G.; Mohr, L.; Lopata-Finch, E.; Emani, C.; Rathore, K.S. Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol. Biol 2002, 50, 463–474. [Google Scholar]
- Boyko, A.; Molinier, J.; Chatter, W.; Laroche, A.; Kovalchuk, I. Acute but not chronic exposure to abiotic stress results in transient reduction of expression levels of the transgene driven by the 35S promoter. New Biotechnol 2010, 27, 70–77. [Google Scholar]
- Yin, T.; Wu, H.; Zhang, S.; Lu, H.; Zhang, L.; Xu, Y.; Chen, D.; Liu, J. Two negative cis-regulatory regions involved in fruit-specific promoter activity from watermelon (Citrullus vulgaris S.). J. Exp. Bot 2009, 60, 169–185. [Google Scholar]
- Wilkie, G.S.; Dickson, K.S.; Gray, N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci 2003, 28, 182–188. [Google Scholar]
- Tran, M.K.; Schultz, C.J.; Baumann, U. Conserved upstream open reading frames in higher plants. BMC Genomics 2008, 9, 361. [Google Scholar]
- Wu, J.; Miller, B.L. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell Biol 1997, 17, 6191–6201. [Google Scholar]
- Zheng, X.; Deng, W.; Luo, K.; Duan, H.; Chen, Y.; McAvoy, R.; Song, S.; Pei, Y.; Li, Y. The cauliflower mosaic virus (CaMV) 35S promoter sequence alters the level and patterns of activity of adjacent tissue-and organ-specific gene promoters. Plant Cell Rep 2007, 26, 1195–1203. [Google Scholar]
- Yang, Y.; Singer, S.D.; Liu, Z. Evaluation and comparison of the insulation efficiency of three enhancer-blocking insulators in plants. Plant Cell Tissue Organ Cult 2011, 105, 405–414. [Google Scholar]
- Qi, R.F.; Song, Z.W.; Chi, C.W. Structural features and molecular evolution of Bowman-Birk protease inhibitors and their potential application. Acta Biochim. Biophys. Sin 2005, 37, 283–292. [Google Scholar]
- Shitan, N.; Horiuchi, K.; Sato, F.; Yazaki, K. Bowman-birk proteinase inhibitor confers heavy metal and multiple drug tolerance in yeast. Plant Cell Physiol 2007, 48, 193–197. [Google Scholar]
- Qu, L.J.; Chen, J.; Liu, M.; Pan, N.; Okamoto, H.; Lin, Z.; Li, C.; Li, D.; Wang, J.; Zhu, G.; et al. Molecular cloning and functional analysis of a novel type of Bowman-Birk inhibitor gene family in rice. Plant Physiol 2003, 133, 560–570. [Google Scholar]
- Eckelkamp, C.; Ehmann, B.; Schopfer, P. Wound-induced systemic accumulation of a transcript coding for a Bowman-Birk trypsin inhibitor-related protein in maize (Zea mays L.) seedlings. FEBS Lett 1993, 323, 73–76. [Google Scholar]
- Rohrmeier, T.; Lehle, L. WIP1, a wound-inducible gene from maize with homology to Bowman-Birk proteinase inhibitors. Plant Mol. Biol 1993, 22, 783–792. [Google Scholar]
- Tiffin, P.; Gaut, B.S. Molecular evolution of the wound-induced serine protease inhibitor wip1 in Zea and related genera. Mol. Biol. Evol 2001, 18, 2092–2101. [Google Scholar]
- Maize Genetics and Genomics Database. Available online: http://www.maizegdb.org/ (accessed on 20 May 2010).
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002, 30, 325–327. [Google Scholar]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 1999, 27, 297–300. [Google Scholar]
- Reese, M.G. Neural Network Promoter Prediction. Available online: http://www.fruitfly.org/seq_tools/promoter.html (accessed on 27 October 2010).
- Walker-Simmons, M.; Holländer-Czytko, H.; Andersen, J.K.; Ryan, C.A. Wound signals in plants: A systemic plant wound signal alters plasma membrane integrity. Proc. Natl. Acad. Sci. USA 1984, 81, 3737–3741. [Google Scholar]
- An, G.; Mitra, A.; Choi, H.K.; Costa, M.A.; An, K.; Thornburg, R.W.; Ryan, C.A. Functional analysis of the 3′control region of the potato wound-inducible proteinase inhibitor II gene. Plant Cell 1989, 1, 115–122. [Google Scholar]
- Xu, D.; McElroy, D.; Thornburg, R.W.; Wu, R. Systemic induction of a potato pin2 promoter by wounding, methyl iasmonate, and abscisic acid in transgenic rice plants. Plant Mol. Biol 1993, 22, 573–588. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar]
- Yoo, S.Y.; Bomblies, K.; Yoo, S.K.; Yang, J.W.; Choi, M.S.; Lee, J.S.; Weigel, D.; Ahn, J.H. The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 2005, 221, 523–530. [Google Scholar]
- Singer, S.D.; Cox, K.D.; Liu, Z. Both the constitutive cauliflower mosaic virus 35S and tissue-specific AGAMOUS enhancers activate transcription autonomously in Arabidopsis thaliana. Plant Mol. Biol 2010, 74, 293–305. [Google Scholar]
- Chen, X.; Wang, Z.; Gu, R.; Fu, J.; Wang, J.; Zhang, Y.; Wang, M.; Zhang, J.; Jia, J.; Wang, G. Isolation of the maize Zpu1 gene promoter and its functional analysis in transgenic tobacco plants. Plant Cell Rep 2007, 26, 1555–1565. [Google Scholar]
- Lü, S.; Gu, H.; Yuan, X.; Wang, X.; Wu, A.M.; Qu, L.; Liu, J.Y. The GUS reporter-aided analysis of the promoter activities of a rice metallothionein gene reveals different regulatory regions responsible for tissue-specific and inducible expression in transgenic Arabidopsis. Transgenic Res 2007, 16, 177–191. [Google Scholar]
- Ren, Y.; Zhao, J. Functional analysis of the rice metallothionein gene OsMT2b promoter in transgenic Arabidopsis plants and rice germinated embryos. Plant Sci 2009, 176, 528–538. [Google Scholar]
- Wilmink, A.; van de Ven, B.C.; Dons, J.J. Activity of constitutive promoters in various species from the Liliaceae. Plant Mol. Biol 1995, 28, 949–955. [Google Scholar]
- Finkelstein, R.; Lynch, T.; Reeves, W.; Petitfils, M.; Mostachetti, M. Accumulation of the transcription factor ABA-insensitive (ABI)4 is tightly regulated post-transcriptionally. J. Exp. Bot 2011, 62, 3971–3979. [Google Scholar]
- Park, S.H.; Yi, N.; Kim, Y.S.; Jeong, M.H.; Bang, S.W.; Choi, Y.D.; Kim, J.K. Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot 2010, 61, 2459–2467. [Google Scholar]
- Caspar, T.; Quail, P.H. Promoter and leader regions involved in the expression of the Arabidopsis ferredoxin A gene. Plant J 1993, 3, 161–174. [Google Scholar]
- Gallie, D.R.; Sleat, D.E.; Watts, J.W.; Turner, P.C.; Wilson, T.M.A. Mutational analysis of the tobacco mosaic virus 5′-leader for altered ability to enhance translation. Nucleic Acids Res 1988, 16, 883–893. [Google Scholar]
- Gallie, D.R.; Walbot, V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res 1992, 20, 4631–4638. [Google Scholar]
- De Amicis, F.; Patti, T.; Marchetti, S. Improvement of the pBI121 plant expression vector by leader replacement with a sequence combining a poly (CAA) and a CT motif. Transgenic Res 2007, 16, 731–738. [Google Scholar]
- Morris, D.R.; Geballe, A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol 2000, 20, 8635–8642. [Google Scholar]
- Singer, S.D.; Cox, K.D.; Liu, Z. Enhancer-promoter interference and its prevention in transgenic plants. Plant Cell Rep 2011, 30, 723–731. [Google Scholar]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res 1997, 25, 4692–4693. [Google Scholar]
- Horsch, R.; Fry, J.; Hoffman, N.; Eichholz, D.; Rogers, S.; Fraley, R. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998, 16, 735–743. [Google Scholar]
- Goto, F.; Yoshihara, T.; Shigemoto, N.; Toki, S.; Takaiwa, F. Iron fortification of rice seed by the soybean ferritin gene. Nat. Biotechnol 1999, 17, 282–286. [Google Scholar]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 1987, 6, 3901–3907. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]








| Primers | Sequence |
|---|---|
| F1737 | 5′-AACTGCAGGGCTCCGTTCTACTTGACT-3′ |
| R1737 | 5′-CGGGATCCGGTCTCGGACGAGCTGTTCTT-3′ |
| F1231 | 5′-AACTGCAGTTTGTTTTGGATTATAAT-3′ |
| F1191 | 5′-AACTGCAGCGCGTCGCCGTCTCTTACTGT-3′ |
| F791 | 5′-AACTGCAGTTCTTTTTAGTTGTCGCTGGA-3′ |
| F491 | 5′-AACTGCAGGGGAAGATTATTACTATACAC-3′ |
| R1231 | 5′-CGGGATCCTTCCTCGTGATCATTTCCG-3′ |
| F1231A | 5′-AACTGCAGCCTTACATAGTGGTTGGT-3′ |
| F1231B | 5′-AACTGCAGGCTTCTATTTTATCACAC-3′ |
| F1231C | 5′-AACTGCAGGGCGTCCATTTGTATCTC-3′ |
| R1231a | 5′-CGGGATCCATGGATGGATAGACGCTT-3′ |
| R1231b | 5′-CGGGATCCGCCGAGATCTGAACATGC-3′ |
| R1231c | 5′-CGGGATCCCCTCTCGTAGAAGCAAGC-3′ |
| The amplified fragment | Forward Primer | Reverse Primer |
|---|---|---|
| Wip1500 | F1231 | R1737 |
| Wip1231 | F1231 | R1231 |
| Wip1191 | F1191 | R1737 |
| Wip791 | F791 | R1737 |
| Wip491 | F491 | R1737 |
| Wip1231A | F1231A | R1231 |
| Wip931 | F1191 | R1231 |
| Wip1231C | F1231C | R1231 |
| Wip1231a | F1231 | R1231a |
| Wip1231b | F1231 | R1231b |
| Wip1231c | F1231 | R1231c |
| Wip1231Aa | F1231A | R1231a |
| Wip1231Ab | F1231A | R1231b |
| Wip1231Ac | F1231A | R1231c |
| Wip1231Ba | F1231B | R1231a |
| Wip1231Bb | F1231B | R1231b |
| Wip1231Bc | F1231B | R1231c |
| Wip1231Ca | F1231C | R1231a |
| Wip1231Cb | F1231C | R1231b |
| Wip1231Cc | F1231C | R1231c |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhang, S.; Lian, Y.; Liu, Y.; Wang, X.; Liu, Y.; Wang, G. Characterization of a Maize Wip1 Promoter in Transgenic Plants. Int. J. Mol. Sci. 2013, 14, 23872-23892. https://doi.org/10.3390/ijms141223872
Zhang S, Lian Y, Liu Y, Wang X, Liu Y, Wang G. Characterization of a Maize Wip1 Promoter in Transgenic Plants. International Journal of Molecular Sciences. 2013; 14(12):23872-23892. https://doi.org/10.3390/ijms141223872
Chicago/Turabian StyleZhang, Shengxue, Yun Lian, Yan Liu, Xiaoqing Wang, Yunjun Liu, and Guoying Wang. 2013. "Characterization of a Maize Wip1 Promoter in Transgenic Plants" International Journal of Molecular Sciences 14, no. 12: 23872-23892. https://doi.org/10.3390/ijms141223872




