Transcriptional Analysis of Hair Follicle-Derived Keratinocytes from Donors with Atopic Dermatitis Reveals Enhanced Induction of IL32 Gene by IFN-γ
Abstract
:1. Introduction
2. Results and Discussion
2.1. Gene Expression Profile of FDKs
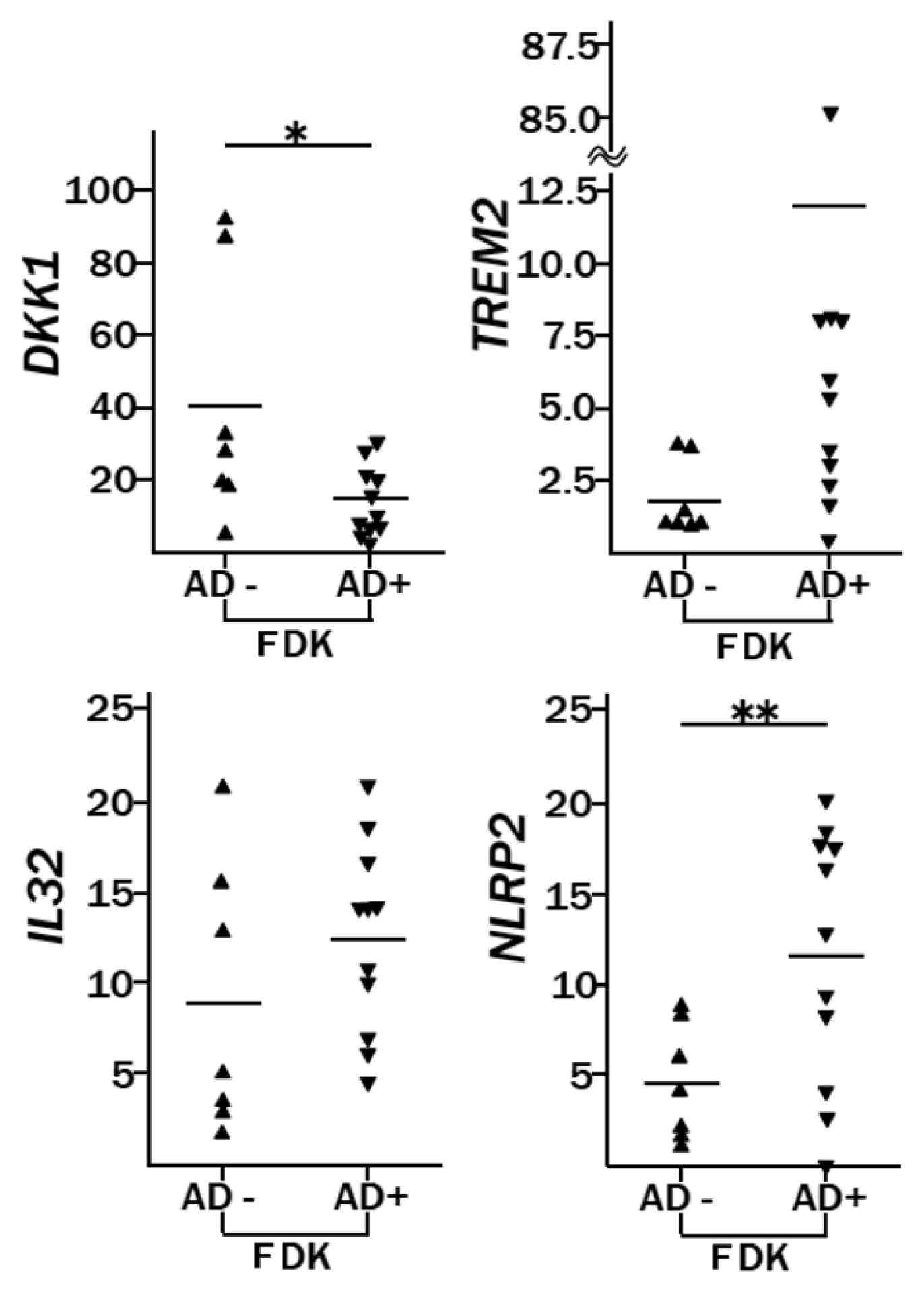
2.2. Genes Differentially Expressed between AD-FDKs and Non-AD-FDKs
2.3. Enhanced Induction of IL32 and IL1B by IFN-γ Treatment in AD-FDKs Compared to Non-AD-FDKs
3. Experimental Section
3.1. Plucked Hair
3.2. Cell Culture and RNA Extraction
3.3. Microarray Analysis
3.4. Real-Time RT-PCR
3.5. Statistical Analysis
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Cotsarelis, G.; Sun, T.T.; Lavker, R.M. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 1990, 61, 1329–1337. [Google Scholar]
- Taylor, G.; Lehrer, M.S.; Jensen, P.J.; Sun, T.T.; Lavker, R.M. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 2000, 102, 451–461. [Google Scholar]
- Watt, F.M.; Hogan, B.L. Out of Eden: Stem cells and their niches. Science 2000, 287, 1427–1430. [Google Scholar]
- Tumbar, T.; Guasch, G.; Greco, V.; Blanpain, C.; Lowry, W.E.; Rendl, M.; Fuchs, E. Defining the epithelial stem cell niche in skin. Science 2004, 303, 359–363. [Google Scholar]
- Ito, M.; Liu, Y.; Yang, Z.; Nguyen, J.; Liang, F.; Morris, R.J.; Cotsarelis, G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med 2005, 11, 1351–1354. [Google Scholar]
- Cotsarelis, G. Epithelial stem cells: A folliculocentric view. J. Invest. Dermatol 2006, 126, 1459–1468. [Google Scholar]
- Sasahara, Y.; Yoshikawa, Y.; Morinaga, T.; Nakano, Y.; Kanazawa, N.; Kotani, J.; Kawamata, S.; Murakami, Y.; Takeuchi, K.; Inoue, C.; et al. Human keratinocytes derived from the bulge region of hair follicles are refractory to differentiation. Int. J. Oncol 2009, 34, 1191–1199. [Google Scholar]
- Eckert, R.L.; Adhikary, G.; Balasubramanian, S.; Rorke, E.A.; Vemuri, M.C.; Boucher, S.E.; Bickenbach, J.R.; Kerr, C. Biochemistry of epidermal stem cells. Biochim. Biophys. Acta 2012, 1830, 2427–2434. [Google Scholar]
- Beck, B.; Blanpain, C. Mechanisms regulating epidermal stem cells. EMBO J 2012, 31, 2067–2075. [Google Scholar]
- Yoshikawa, Y.; Sasahara, Y.; Kitano, Y.; Kanazawa, N.; Shima, H.; Hashimoto-Tamaoki, T. Upregulation of genes orchestrating keratinocyte differentiation, including the novel marker gene ID2, by contact sensitizers in human bulge-derived keratinocytes. J. Biochem. Mol. Toxicol 2010, 24, 10–20. [Google Scholar]
- Brandt, E.B.; Sivaprasad, U. Th2 cytokines and atopic dermatitis. J. Clin. Cell. Immunol 2011, 2, 110. [Google Scholar]
- De Benedetto, A.; Agnihothri, R.; McGirt, L.Y.; Bankova, L.G.; Beck, L.A. Atopic dermatitis: A disease caused by innate immune defects? J. Invest. Dermatol 2009, 129, 14–30. [Google Scholar]
- Palmer, C.N.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.; et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet 2006, 38, 441–446. [Google Scholar]
- Hubiche, T.; Ged, C.; Benard, A.; Leaute-Labreze, C.; McElreavey, K.; de Verneuil, H.; Taieb, A.; Boralevi, F. Analysis of SPINK 5, KLK 7 and FLG genotypes in a French atopic dermatitis cohort. Acta Derm. Venereol 2007, 87, 499–505. [Google Scholar]
- Thyssen, J.; Carlsen, B.; Bisgaard, H.; Giwercman, C.; Johansen, J.; Linneberg, A.; Meldgaard, M.; Szecsi, P.; Stender, S.; Menne, T. Individuals who are homozygous for the 2282del4 and R501X filaggrin null mutations do not always develop dermatitis and complete long-term remission is possible. J. Eur. Acad Dermatol. Venereol 2012, 26, 386–389. [Google Scholar]
- Wittmann, M.; Werfel, T. Interaction of keratinocytes with infiltrating lymphocytes in allergic eczematous skin diseases. Curr. Opin. Allergy Clin. Immunol 2006, 6, 329–334. [Google Scholar]
- Albanesi, C. Keratinocytes in allergic skin diseases. Curr. Opin. Allergy Clin. Immunol 2010, 10, 452–426. [Google Scholar]
- Trautmann, A.; Akdis, M.; Kleemann, D.; Altznauer, F.; Simon, H.U.; Graeve, T.; Noll, M.; Brocker, E.B.; Blaser, K.; Akdis, C.A. T cell-mediated Fas-induced keratinocyte apoptosis plays a key pathogenetic role in eczematous dermatitis. J. Clin. Invest 2000, 106, 25–35. [Google Scholar]
- Rebane, A.; Zimmermann, M.; Aab, A.; Baurecht, H.; Koreck, A.; Karelson, M.; Abram, K.; Metsalu, T.; Pihlap, M.; Meyer, N.; et al. Mechanisms of IFN-gamma-induced apoptosis of human skin keratinocytes in patients with atopic dermatitis. J. Allergy Clin. Immunol 2012, 129, 1297–1306. [Google Scholar]
- Meyer, N.; Zimmermann, M.; Bürgler, S.; Bassin, C.; Woehrl, S.; Moritz, K.; Rhyner, C.; Indermitte, P.; Schmid-Grendelmeier, P.; Akdis, M.; et al. IL-32 is expressed by human primary keratinocytes and modulates keratinocyte apoptosis in atopic dermatitis. J. Allergy Clin. Immunol 2010, 125, 858–865. [Google Scholar]
- Bernard, F.X.; Pedretti, N.; Rosdy, M.; Deguercy, A. Comparison of gene expression profiles in human keratinocyte mono-layer cultures, reconstituted epidermis and normal human skin; transcriptional effects of retinoid treatments in reconstituted human epidermis. Exp. Dermatol 2002, 11, 59–74. [Google Scholar]
- Amano, S.; Akutsu, N.; Ogura, Y.; Nishiyama, T. Increase of laminin 5 synthesis in human keratinocytes by acute wound fluid, inflammatory cytokines and growth factors, and lysophospholipids. Br. J. Dermatol 2004, 151, 961–970. [Google Scholar]
- Penas, P.F.; Garcia-Diez, A.; Sanchez-Madrid, F.; Yanez-Mo, M. Tetraspanins are localized at motility-related structures and involved in normal human keratinocyte wound healing migration. J. Invest. Dermatol 2000, 114, 1126–1135. [Google Scholar]
- Peng, W.M.; Yu, C.F.; Kolanus, W.; Mazzocca, A.; Bieber, T.; Kraft, S.; Novak, N. Tetraspanins CD9 and CD81 are molecular partners of trimeric FcvarepsilonRI on human antigen-presenting cells. Allergy 2011, 66, 605–611. [Google Scholar]
- Gudjonsson, J.E.; Johnston, A.; Stoll, S.W.; Riblett, M.B.; Xing, X.; Kochkodan, J.J.; Ding, J.; Nair, R.P.; Aphale, A.; Voorhees, J.J.; et al. Evidence for altered Wnt signaling in psoriatic skin. J. Invest. Dermatol 2010, 130, 1849–1859. [Google Scholar]
- Yamaguchi, Y.; Passeron, T.; Hoashi, T.; Watabe, H.; Rouzaud, F.; Yasumoto, K.; Hara, T.; Tohyama, C.; Katayama, I.; Miki, T.; et al. Dickkopf 1 (DKK1) regulates skin pigmentation and thickness by affecting Wnt/beta-catenin signaling in keratinocytes. FASEB. J 2008, 22, 1009–1020. [Google Scholar]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar]
- Bruey, J.M.; Bruey-Sedano, N.; Newman, R.; Chandler, S.; Stehlik, C.; Reed, J.C. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-kappaB and caspase-1 activation in macrophages. J. Biol. Chem 2004, 279, 51897–51907. [Google Scholar]
- Martinon, F.; Tschopp, J. Inflammatory caspases: Linking an intracellular innate immune system to autoinflammatory diseases. Cell 2004, 117, 561–574. [Google Scholar]
- Nagao, K.; Kobayashi, T.; Moro, K.; Ohyama, M.; Adachi, T.; Kitashima, D.Y.; Ueha, S.; Horiuchi, K.; Tanizaki, H.; Kabashima, K.; et al. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol 2012, 13, 744–752. [Google Scholar]


| Function annotation | p value | Predicted activation state in FDKs | Regulation z-score |
|---|---|---|---|
| Migration of cells | 6.58 × 10−7 | Increased | 3.025 |
| Cell movement | 1.77 × 10−6 | Increased | 2.942 |
| Adhesion of immune cells | 8.47 × 10−4 | Increased | 2.893 |
| Cell spreading | 5.16 × 10−5 | Increased | 2.857 |
| Transmigration of cells | 4.98 × 10−6 | Increased | 2.635 |
| Binding of cells | 1.93 × 10−3 | Increased | 2.438 |
| MAPKKK cascade | 1.80 × 10−3 | Increased | 2.387 |
| Expression of RNA | 1.88 × 10−3 | Increased | 2.298 |
| Autophagy | 1.41 × 10−4 | Increased | 2.26 |
| Regression of embryonic tissue | 2.10 × 10−3 | Increased | 2.2 |
| Initiation of cell death | 2.04 × 10−3 | Increased | 2 |
| Alignment of chromosomes | 1.60 × 10−6 | Decreased | −2.608 |
| Function annotation | p value | Predicted activation state | Regulation z-score |
|---|---|---|---|
| Cellular response to therapeutics: sensitivity of cells | 0.0335 | Increased | 2.543 |
| Molecular transport: concentration of glutathione | 0.0126 | Increased | 2.416 |
| Cell death | 0.0001 | Increased | 2.409 |
| Apoptosis | 0.0078 | Increased | 2.048 |
| Cell proliferation | 0.0058 | Decreased | −3.071 |
Supplementary Files
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yoshikawa, Y.; Sasahara, Y.; Takeuchi, K.; Tsujimoto, Y.; Hashida-Okado, T.; Kitano, Y.; Hashimoto-Tamaoki, T. Transcriptional Analysis of Hair Follicle-Derived Keratinocytes from Donors with Atopic Dermatitis Reveals Enhanced Induction of IL32 Gene by IFN-γ. Int. J. Mol. Sci. 2013, 14, 3215-3227. https://doi.org/10.3390/ijms14023215
Yoshikawa Y, Sasahara Y, Takeuchi K, Tsujimoto Y, Hashida-Okado T, Kitano Y, Hashimoto-Tamaoki T. Transcriptional Analysis of Hair Follicle-Derived Keratinocytes from Donors with Atopic Dermatitis Reveals Enhanced Induction of IL32 Gene by IFN-γ. International Journal of Molecular Sciences. 2013; 14(2):3215-3227. https://doi.org/10.3390/ijms14023215
Chicago/Turabian StyleYoshikawa, Yoshie, Yusuke Sasahara, Katsuyuki Takeuchi, Yoshimasa Tsujimoto, Takashi Hashida-Okado, Yukio Kitano, and Tomoko Hashimoto-Tamaoki. 2013. "Transcriptional Analysis of Hair Follicle-Derived Keratinocytes from Donors with Atopic Dermatitis Reveals Enhanced Induction of IL32 Gene by IFN-γ" International Journal of Molecular Sciences 14, no. 2: 3215-3227. https://doi.org/10.3390/ijms14023215




