Ribosomal Initiation Complex Assembly within the Wild-Strain of Coxsackievirus B3 and Live-Attenuated Sabin3-like IRESes during the Initiation of Translation
Abstract
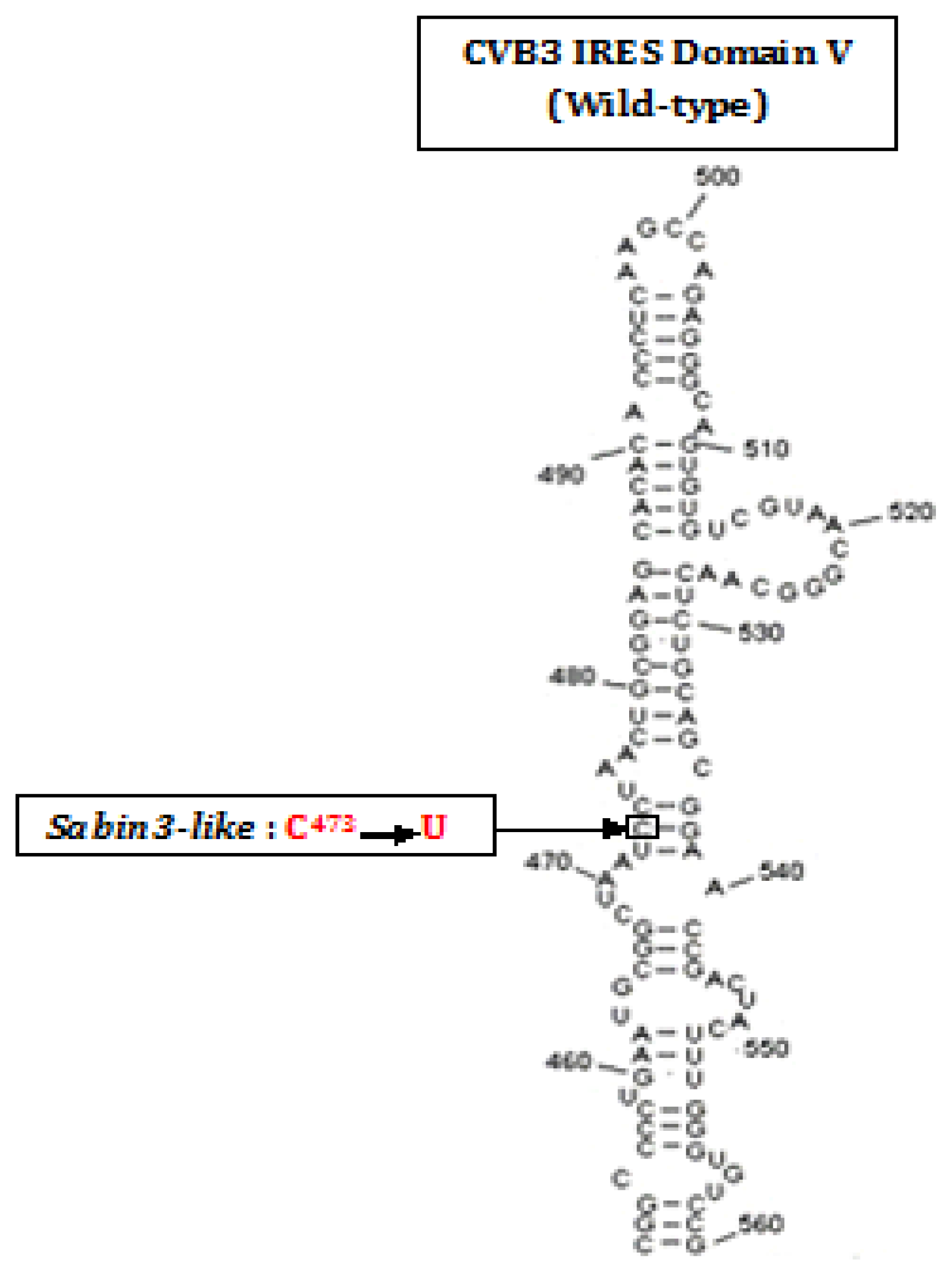
:1. Introduction
2. Results
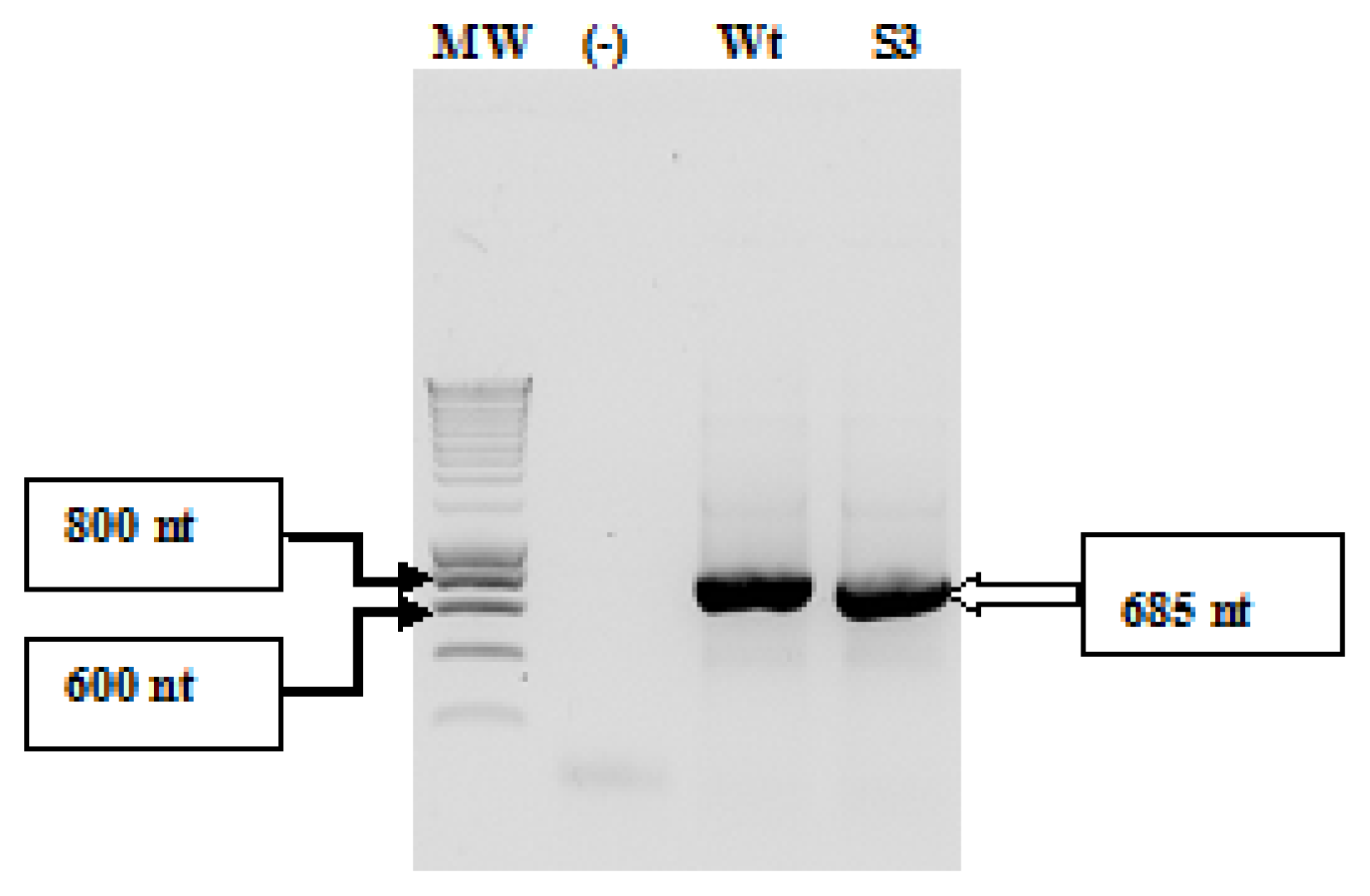
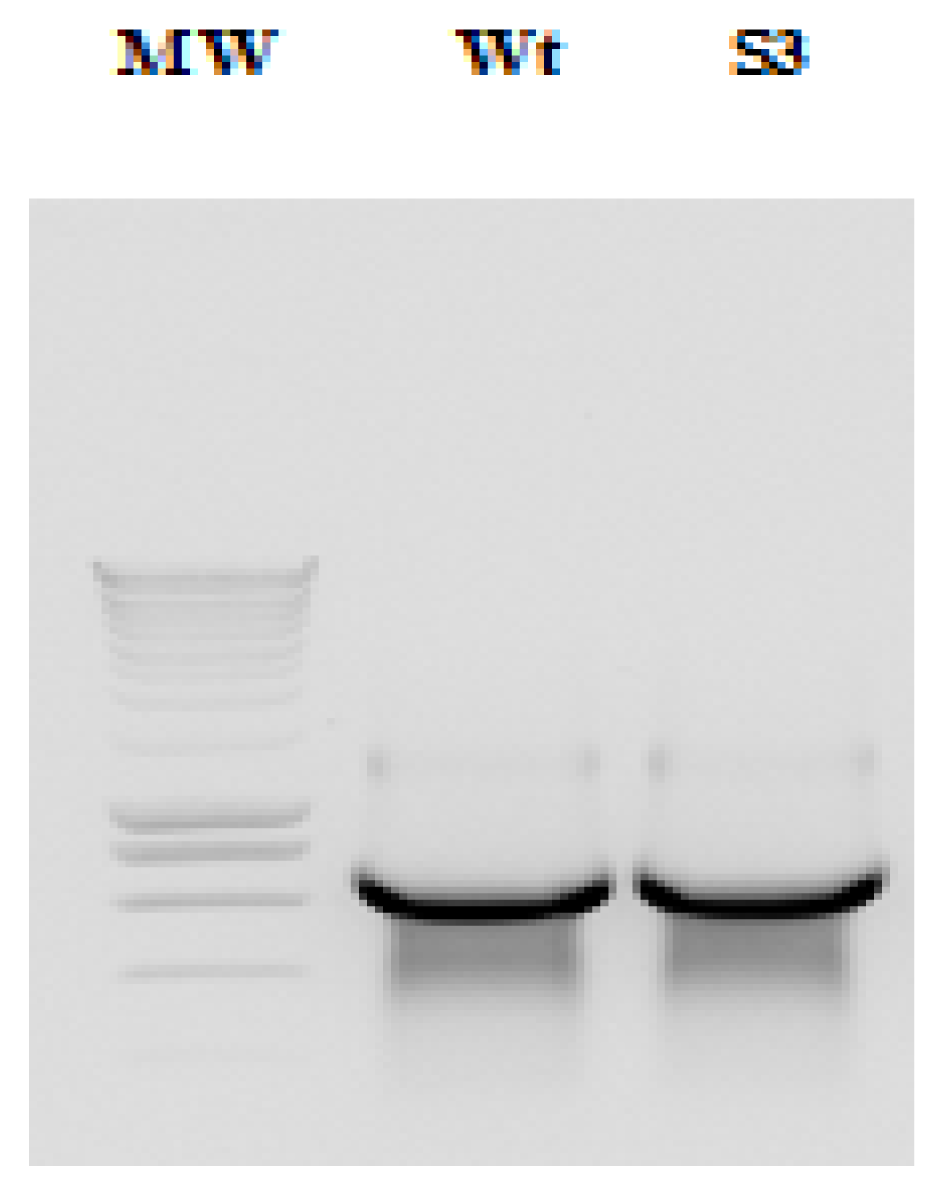
2.1. Amplification and Cloning of the IRES
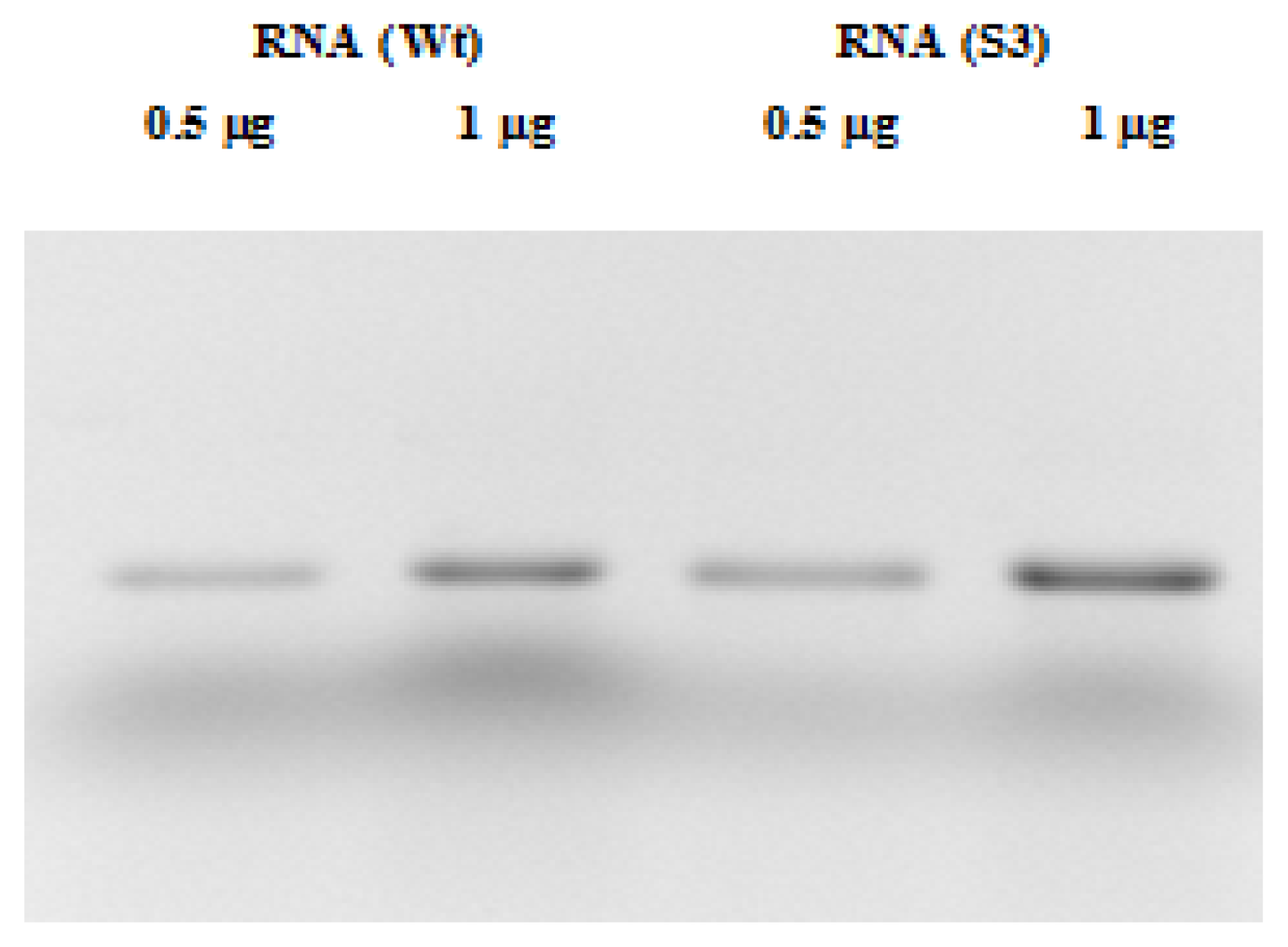
2.2. RNA and Fluorescent Labeling
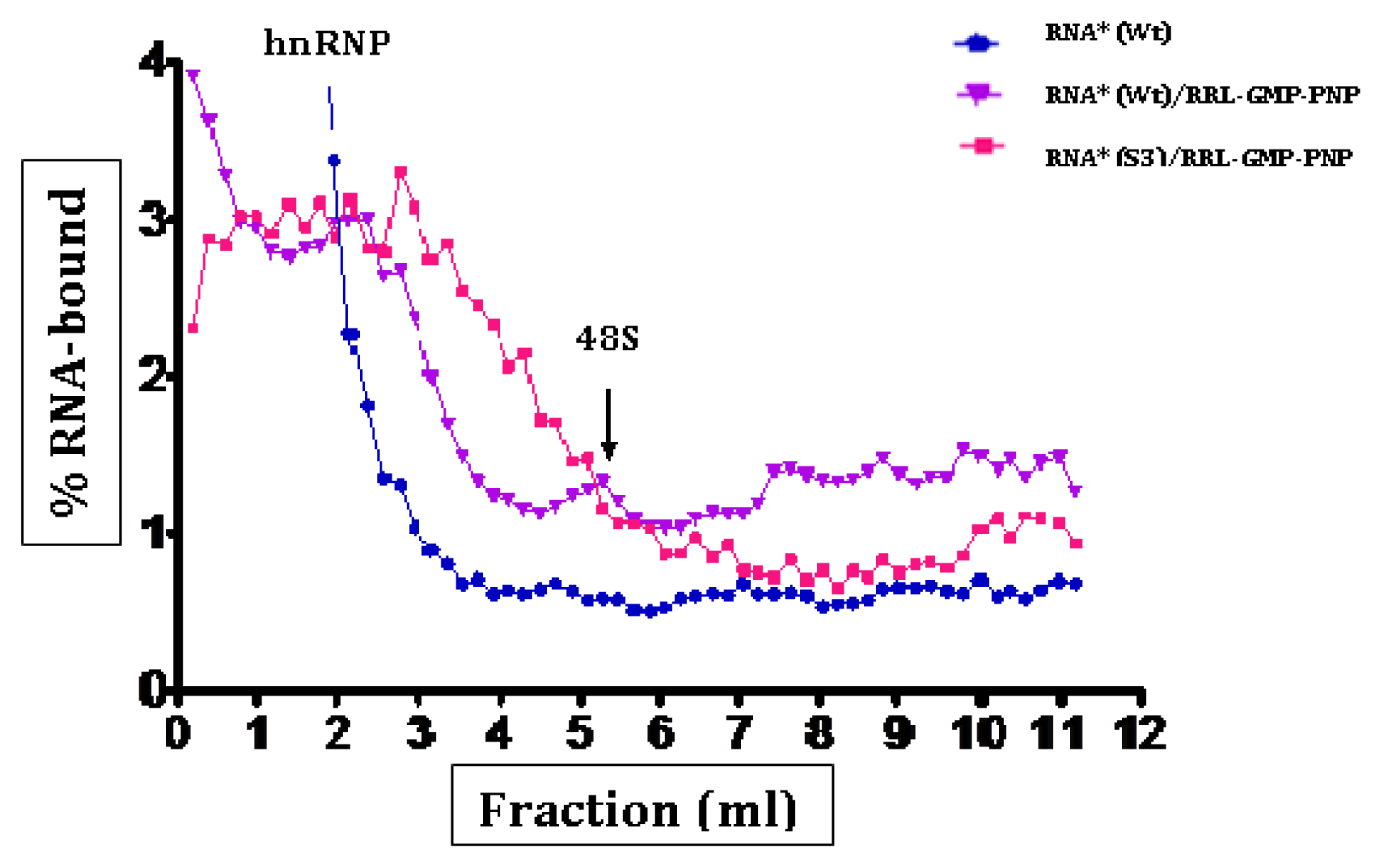
2.3. Initiation Complexes Assembly
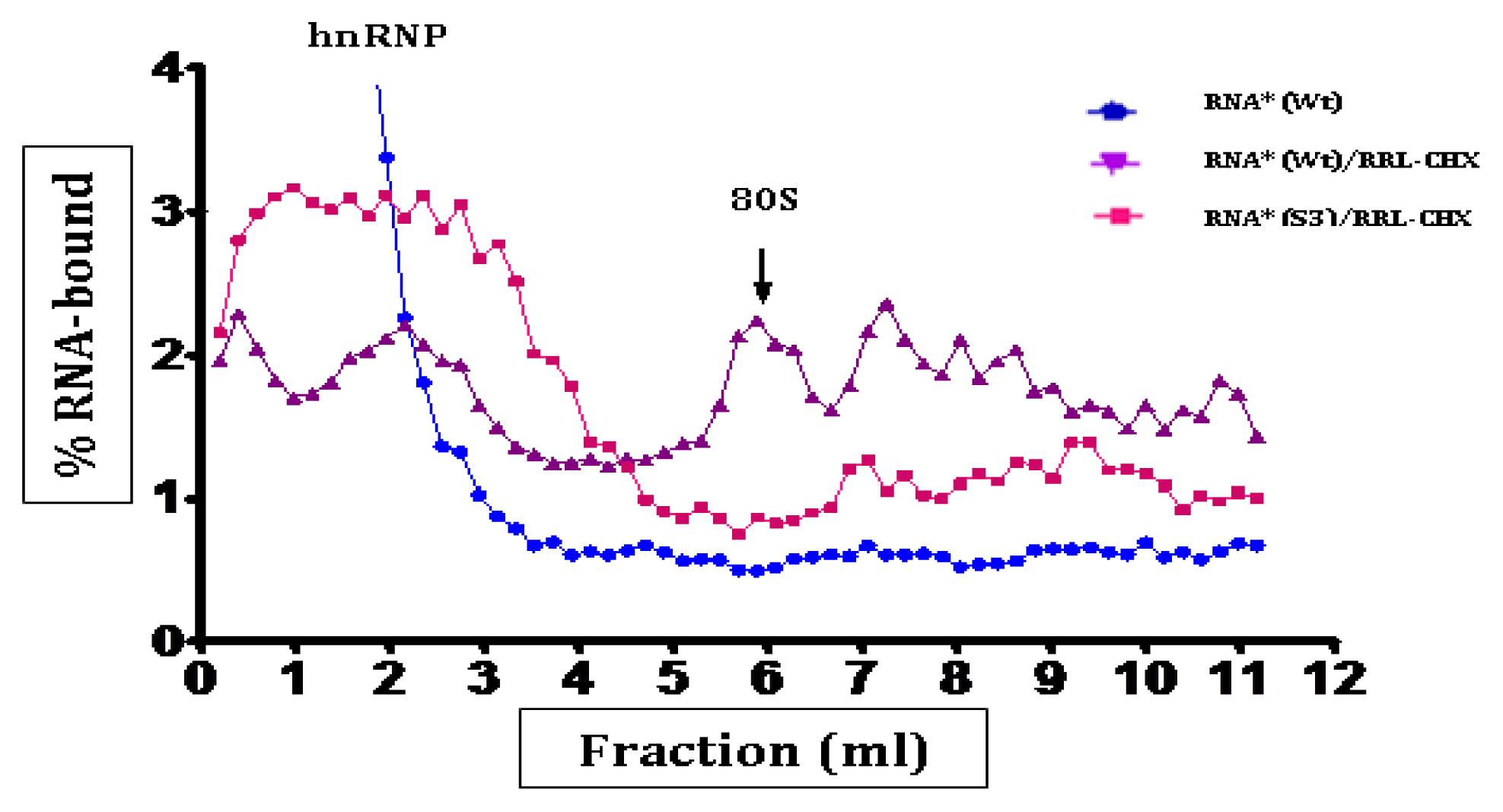
3. Discussion
4. Experimental Section
4.1. Virus
4.2. Plasmids and Cloning Conditions
4.3. Bacterial Strains
4.4. Viral RNA Extraction
4.5. Reverse Transcription (RT)
4.6. Polymerase Chain Reaction
4.7. Cloning of the IRES in the pUC19 Vector
4.8. Screening of Recombinant Clones
4.9. RNA and in Vitro Transcription
4.10. Periodate Oxidation and Fluorophore Coupling
4.11. Mobility of Initiation Complexes during Sucrose Density Gradients Centrifugation
5. Conclusion
Acknowledgments
Conflict of Interest
References
- Kishimoto, C.; Hiraoka, Y. Clinical and experimental studies in myocarditis. Curr. Opin. Cardiol 1994, 9, 349–356. [Google Scholar]
- Reyes, M.P.; Lerner, A.M. Coxsackievirus myocarditis with special reference to acute and chronic effects. Prog. Cardiovasc. Dis 1985, 27, 373–394. [Google Scholar]
- Woodruff, J.F. Viral myocarditis. A review. Am. J. Pathol 1980, 101, 425–484. [Google Scholar]
- Arnesjo, B.; Eden, T.; Ihse, I.; Nordenfelt, E.; Ursing, B. Enterovirus infections in acute pancreatitis. Scand. J. Gastroenterol 1976, 11, 645–649. [Google Scholar]
- Imrie, C.W.; Ferguson, J.C.; Sommerville, R.G. Coxsackie and mumpsvirus infection in a prospective study of acute pancreatitis. Gut 1977, 18, 53–56. [Google Scholar]
- Mena, I.; Fischer, C.; Gebhard, J.R.; Perry, C.M.; Harkins, S.; Whitton, J.L. Coxsackievirus infection of the pancreas: Evaluation of receptor expression, pathogenesis, and immunopathology. Virology 2000, 271, 276–288. [Google Scholar]
- Ramsingh, A.I. Coxsackieviruses and pancreatitis. Front. Biosci 1997, 2, e53–e62. [Google Scholar]
- Daley, A.J.; Isaacs, D.; Dwyer, D.E.; Gilbert, G.L. A cluster of cases of neonatal coxsackievirus B meningitis and myocarditis. J. Pediatr. Child. Health 1998, 34, 196–198. [Google Scholar]
- Modlin, J.F.; Rotbart, H.A. Group B coxsackie disease in children. Curr. Top. Microbiol. Immunol 1997, 223, 53–80. [Google Scholar]
- Beck, M.A.; Chapman, N.M.; McManus, B.M.; Mullican, J.C.; Tracy, S. Secondary enterovirus infection in the murine model of myocarditis. Pathologic and immunologic aspects. Am. J. Pathol 1990, 136, 669–681. [Google Scholar]
- Esfandiarei, M.; McManus, B.M. Molecular biology and pathogenesis of viral myocarditis. Annu. Rev. Pathol 2008, 3, 127–155. [Google Scholar]
- Kim, K.S.; Hufnagel, G.; Chapman, N.M.; Tracy, S. The group B coxsackieviruses and myocarditis. Rev. Med. Virol 2001, 11, 355–368. [Google Scholar]
- Tu, Z.; Chapman, N.; Hufnagel, G.; Tracy, S.; Romero, J.R.; Barry, W.H.; Zhao, L.; Currey, K.; Shapiro, B. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the Genomic 5′ Nontranslated region. J. Virol 1995, 69, 4607–4618. [Google Scholar]
- Tracy, S.; Chapman, N.M.; Tu, Z. Coxsackievirus B3 from an infectious cDNA copy of the genome is cardiovirulent in mice. Arch. Virol 1992, 122, 399–409. [Google Scholar]
- See, D.M.; Tilles, J.G. Efficacy of a polyvalent inactivated-virus vaccine in protecting mice from infection with clinical strains of group B coxsackieviruses. Scand. J. Infect. Dis 1994, 26, 739–747. [Google Scholar]
- Ben M’hadheb-Gharbi, M.; Gharbi, J.; Paulous, S.; Brocard, M.; Komaromva, A.; Aouni, M.; Kean, K.M. Effects of the Sabin-like mutations in domain V of the internal ribosome entry segment on translational efficiency of the Coxsackievirus B3. Mol. Gen. Genomics 2006, 276, 402–412. [Google Scholar]
- Ben M’hadheb-Gharbi, M.; Kean, K.M.; Gharbi, J. Molecular analysis of the role of IRES stem-loop V in replicative capacities and translation efficiencies of Coxsackievirus B3 mutants. Mol. Biol. Rep 2009, 36, 255–262. [Google Scholar]
- Dan, M.; Chantler, J.K. A genetically engineered attenuated coxsackievirus B3 strain protects mice against lethal infection. J. Virol 2005, 79, 9285–9295. [Google Scholar]
- Fohlman, J.; Pauksen, K.; Morein, B.; Bjare, U.; Ilbäck, N.G.; Friman, G. High yield production of an inactivated coxsackie B3 adjuvant vaccine with protective effect against experimental myocarditis. Scand. J. Infect. Dis. Suppl 1993, 88, 103–108. [Google Scholar]
- Gauntt, C.J.; Paque, R.E.; Trousdale, M.D.; Gudvangen, R.J.; Barr, D.T.; Lipotich, G.J.; Nealon, T.J.; Duffey, P.S. Temperature-sensitive mutant of coxsackievirus B3 establishes resistance in neonatal mice that protects them during adolescence against coxsackievirus B3-induced myocarditis. Infect. Immun 1983, 39, 851–864. [Google Scholar]
- Henke, A.; Wagner, E.; Whitton, J.L.; Zell, R.; Stelzner, A. Protection of mice against lethal coxsackievirus B3 infection by using DNA immunization. J. Virol 1998, 72, 8327–8331. [Google Scholar]
- Hunziker, I.P.; Cornell, C.T.; Whitton, J.L. Deletions within the 5′UTR of coxsackievirus B3: Consequences for virus translation and replication. Virology 2007, 360, 120–128. [Google Scholar]
- Bailey, J.M.; Tapprich, W.E. Structure of the 5′-Nontranslated Region of the coxsackievirus B3 genome: Chemical modification and comparative sequence analysis. J. Virol 2007, 81, 650–668. [Google Scholar]
- Yang, D.; Wilson, J.E.; Anderson, D.R.; Bohunek, L.; Cordeiro, C.; Kandol, R.; Mcmanus, B.M. In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5′ untranslated region of coxsackievirus B3: Potential targets for antiviral action of antisense oligomers. Virology 1997, 228, 63–73. [Google Scholar]
- Ray, P.S.; Das, S. La autoantigen is required for the internal ribosome entry site-mediated translation of Coxsackievirus B3 RNA. Nucleic Acid Res 2002, 30, 4500–4508. [Google Scholar]
- Belsham, G.J.; Sonenberg, N. Picornavirus RNA-translation: Role for cellular proteins. Trends Microbiol 2000, 8, 330–335. [Google Scholar]
- Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol 1996, 16, 6859–6869. [Google Scholar]
- Ventoso, I.; MacMillan, S.E.; Hershey, J.W.; Carrasco, L. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Lett 1998, 435, 79–83. [Google Scholar]
- Jang, S.K.; Krausslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol 1988, 62, 2636–2643. [Google Scholar]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar]
- Bedard, K.M.; Daijogo, S.; Semler, B.L. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J 2007, 26, 459–467. [Google Scholar]
- Jackson, R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans 2005, 33, 1231–1241. [Google Scholar]
- Pestova, T.V.; Kolupaeva, V.G.; Lomakin, I.B.; Pilipenko, E.V.; Shatsky, I.N.; Agol, V.I.; Hellen, C.U.T. Molecular mechanisms of translation initiation in eukaryotes: Colloquium. Proc. Natl. Acad. Sci. USA 2001, 98, 7029–7036. [Google Scholar]
- Hellen, C.U.T.; Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 2001, 15, 1593–1612. [Google Scholar]
- Pestova, T.V.; Lomakin, I.B.; Lee, J.H.; Choi, S.K.; Dever, T.E.; Hellen, C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 2000, 403, 332–335. [Google Scholar]
- Anthony, D.D.; Merrick, W.C. Eukaryotic initiation factor (eIF)-4F. Implications for a role in internal initiation of translation. J. Biol. Chem 1991, 266, 10218–10226. [Google Scholar]
- Ochs, K.; Rust, R.C.; Niepmann, M. Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S Initiation complexes independently of initiator AUG location. J. Virol 1999, 73, 7505–7514. [Google Scholar]
- Pause, A.; Méthot, N.; Svitkin, Y.; Merrick, W.C.; Sonenberg, N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 1994, 13, 1205–1215. [Google Scholar]
- Pestova, T.V.; Shatsky, I.N.; Fletcher, S.P.; Jackson, R.J.; Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 1998, 12, 67–83. [Google Scholar]
- Flanegan, J.B.; Pettersson, R.F.; Ambros, V.; Hewlett, N.J.; Baltimore, D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc. Natl. Acad. Sci. USA 1977, 74, 961–965. [Google Scholar]
- Lee, Y.F.; Nomoto, A.; Detjen, B.M.; Wimmer, E. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 1977, 74, 59–63. [Google Scholar]
- Etchison, D.; Milburn, S.C.; Edery, I.; Sonenberg, N.; Hershey, J.W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220.000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem 1982, 257, 14806–14810. [Google Scholar]
- Krausslich, H.G.; Nicklin, M.J.; Toyoda, H.; Etchison, D.; Wimmer, E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol 1987, 61, 2711–2718. [Google Scholar]
- La Monica, N.; Racaniello, V.R. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol 1989, 63, 2357–2560. [Google Scholar]
- Svitkin, Y.V.; Cammack, N.; Minor, P.D.; Almond, J.W. Translation deficiency of the Sabin type 3 poliovirus genome: Association with an attenuating mutation C472-U. Virology 1990, 175, 103–109. [Google Scholar]
- Malnou, E.C.; Werner, A.; Borman, M.A.; Westhof, E.; Kean, M.K. Effects of vaccine strain mutations in domain V of the internal ribosome entry segment compared in the wild type poliovirus type 1 context. J. Biol. Chem 2004, 279, 10261–10269. [Google Scholar]
- Poehlsgaard, J.; Douthwaite, S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol 2005, 3, 870–881. [Google Scholar]
- Fernandez, J.; Yaman, I.; Huang, C.; Liu, H.; Lopez, A.B.; Komar, A.A.; Caprara, M.G.; Merrick, W.C.; Snider, M.D.; Kaufman, R.J.; et al. Ribosome Stalling Regulates IRES-Mediated Translation in Eukaryotes, a Parallel to Prokaryotic Attenuation. Mol. Cell 2005, 17, 405–416. [Google Scholar]
- Obrig, T.G.; Culp, W.J.; McKeehan, W.L.; Hardesty, B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem 1971, 246, 174–181. [Google Scholar]
- Schneider-Poetsch, T.; Ju, J.; Eyler, E.; Dang, Y.; Bhat, S.; Merrick, W.C.; Green, R.; Shen, B.; Liu, J.O. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol 2010, 6, 209–217. [Google Scholar]
- Robert, F.; Kapp, L.D.; Khan, S.N.; Acker, M.G.; Kolitz, S.; Kazemi, S.; Kaufman, R.J.; Merrick, W.C.; Koromilas, A.E.; Lorsch, J.R.; Pelletier, J. Initiation of protein synthesis by Hepatitis C Virus is refractory to reduced eIF2.GTP.Met-tRNAiMet ternary complex availability. Mol. Biol. Cell 2006, 17, 4632–4644. [Google Scholar]
- Chan, J.; Khan, S.N.; Harvey, I.; Merrick, W.; Pelletier, J. Eukaryotic protein synthesis inhibitors identified by comparison of cytotoxicity profiles. RNA 2004, 10, 528–543. [Google Scholar]
- Kapp, L.D.; Lorsch, J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem 2004, 73, 657–704. [Google Scholar]
- Low, W.K.; Dang, Y.; Schneider-Poetsch, T.; Shi, Z.; Choi, N.S.; Merrick, W.C.; Romo, D.; Liu, J.O. Inhibition of Eukaryotic Translation Initiation by the Marine Natural Product Pateamine A. Mol. Cell 2005, 20, 709–722. [Google Scholar]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol 2010, 11, 113–127. [Google Scholar]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar]
- Fitzgerald, K.D.; Semler, B.L. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochem. Biophys. Acta 2009, 1789, 518–528. [Google Scholar]
- López-Lastra, M.; Rivas, A.; Barria, M.I. Protein synthesis in eukaryotes: The growing biological relevance of cap-independent translation initiation. Biol. Res 2005, 38, 121–146. [Google Scholar]
- Vallejos, M.; Deforges, J.; Plank, T.D.M.; Letelier, A.; Ramdohr, P.; Abraham, C.G.; Valiente-Echeverrìa, F.; Kieft, J.S.; Sargueil, B.; López-Lastra, M. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acid Res 2011, 39, 6186–6200. [Google Scholar]
- Chen, T.C.; Weng, K.F.; Chang, S.C.; Lin, J.Y.; Huang, P.N.; Shih, S.R. Development of antiviral agents for enteroviruses. J. Antimicrob. Chemother 2008, 62, 1169–1173. [Google Scholar]
- Liu, Z.; Carthy, C.M.; Cheung, P.; Bohunek, L.; Wilson, J.E.; McManus, B.M.; Yang, D. Structural and functional analysis of the 5′untranslated region of coxsackievirus B3 RNA: In vivo translational and infectivity studies of full-length mutants. Virology 1999, 265, 206–217. [Google Scholar]
- Ben M’hadheb-Gharbi, M.; Paulous, S.; Aouni, M.; Kean, K.M.; Gharbi, J. The substitution U473→C with Sabin3-like mutation within the IRES attenuate coxsackievirus B3 cardiovirulence. Mol. Biotechnol 2007, 36, 52–60. [Google Scholar]
- Bhattacharyya, S.; Das, S. Mapping of secondary structure of the spacer region within the 5′-untranslated region of the coxsackievirus B3 RNA: Possible role of an apical GAGA loop in binding La protein and influencing internal initiation of translation. Virus Res 2005, 108, 89–100. [Google Scholar]
- Verma, B.; Ponnuswamy, A.; Gnanasundram, S.V.; Das, S. Cryptic AUG is important for 48S ribosomal assembly during internal initiation of translation of coxsackievirus B3 RNA. J. Gen. Virol 2011, 92, 1–10. [Google Scholar]
- King, H.A.; Cobbold, L.C.; Willis, A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans 2010, 38, 1581–1586. [Google Scholar]
- Semler, B.L.; Waterman, M.L. IRES-mediated pathways to polysomes: Nuclear versus cytoplasmic routes. Trends Microbiol 2008, 16, 1–5. [Google Scholar]
- Ochs, K.; Zeller, A.; Saleh, L.; Bassili, G.; Song, Y.; Sonntag, A.; Niepmann, M. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol 2003, 77, 115–122. [Google Scholar]
- Pestova, T.V.; Hellen, C.U.T.; Wimmer, E. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology 1994, 204, 729–737. [Google Scholar]
- Poyry, T.A.; Hentze, M.W.; Jackson, R.J. Construction of regulatable picornavirus IRESes as a test of current models of the mechanism of internal translation initiation. RNA 2001, 7, 647–660. [Google Scholar]
- Iizuka, N.; Yonekawa, H.; Nomoto, A. Nucleotide sequences important for translation initiation of enterovirus RNA. J. Virol 1991, 65, 4867–4873. [Google Scholar]
- Yang, D.; Cheung, P.; Sun, Y.; Yuan, J.; Zhang, H.; Carthy, C.M.; Anderson, D.R.; Bohunek, L.; Wilson, J.E.; Mcmanus, B.M. A Shine-Dalgarno-like sequence mediates in vitro ribosomal internal entry and subsequent scanning for translation initiation of coxsackievirus B3 RNA. Virology 2003, 305, 31–43. [Google Scholar]
- Locker, N.; Easton, L.E.; Lukavsky, P.J. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 2007, 26, 795–805. [Google Scholar]
- Lukavsky, P.J. Structure and function of HCV IRES domains. Virus Res 2009, 139, 166–171. [Google Scholar]
- Pestova, T.V.; de Breyne, S.; Pisarev, A.V.; Abaeva, I.S.; Hellen, C.U. eIF2- dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J 2008, 27, 1060–1072. [Google Scholar]
- Vagner, S.; Galy, B.; Pyronnet, S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep 2001, 10, 893–898. [Google Scholar]
- Verma, B.; Bhattacharyya, S.; Das, S. Polypyrimidine tract binding protein interacts with coxsackievirus B3 RNA and influences its translation. J. Gen. Virol 2010, 91, 1245–1255. [Google Scholar]
- Zell, R.; Ihle, Y.; Seitz, S.; Gündel, U.; Wutzler, P.; Görlach, M. Poly (rC)-binding protein 2 interacts with the oligo (rC) tract of coxsackievirus B3. Biochem. Biophys. Res. Commun 2008, 366, 917–921. [Google Scholar]
- Henis-Korenblit, S.; Shani, G.; Sines, T.; Marash, L.; Shohat, G.; Kimchi, A. The caspase-cleaved DAP5 protein supports internal ribosome entry site mediated translation of death proteins. Proc. Natl. Acad. Sci. USA 2002, 99, 5400–5405. [Google Scholar]
- Skinner, M.; Racaniello, V.; Dunn, G.; Cooper, J.; Minor, P.D.; Almond, J.W. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol 1989, 207, 379–392. [Google Scholar]
- Pilipenko, E.V.; Gmyl, A.P.; Maslova, S.V.; Svitkin, Y.V.; Sinyakov, A.N.; Agol, V.I. Prokaryotic-like cis elements in the cap independent internal initiation of translation on picornavirus RNA. Cell 1992, 68, 119–131. [Google Scholar]
- Bhattacharyya, S.; Verma, B.; Pandey, G.; Das, S. The structure and function of a cis-acting element located upstream of the IRES that influences Coxsackievirus B3 RNA translation. Virology 2008, 377, 345–354. [Google Scholar]
- López de Quinto, S.; Martínez-Salas, E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol 1997, 71, 4171–4175. [Google Scholar]
- Malnou, C.E.; Pöyry, T.A.; Jackson, R.J.; Kean, K.M. Poliovirus internal ribosome entry segment structure alterations that specifically affect function in neuronal cells: Molecular genetic analysis. J. Virol 2002, 76, 10617–10626. [Google Scholar]
- Fernández-Miragall, O.; Martínez-Salas, E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 2003, 9, 1333–1344. [Google Scholar]
- Bhattacharyya, S.; Das, S. An apical GAGA loop within 5′UTR of the coxsackievirus B3 RNA maintains structural organization of the IRES element required for efficient ribosome entry. RNA Biol 2006, 3, 60–68. [Google Scholar]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by Acid Guanidium thiocyanate-phenol-Chloroform extraction. Anal. Biochem 1987, 162, 156–159. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Souii, A.; M'hadheb-Gharbi, M.B.; Sargueil, B.; Brossard, A.; Chamond, N.; Aouni, M.; Gharbi, J. Ribosomal Initiation Complex Assembly within the Wild-Strain of Coxsackievirus B3 and Live-Attenuated Sabin3-like IRESes during the Initiation of Translation. Int. J. Mol. Sci. 2013, 14, 4400-4418. https://doi.org/10.3390/ijms14034400
Souii A, M'hadheb-Gharbi MB, Sargueil B, Brossard A, Chamond N, Aouni M, Gharbi J. Ribosomal Initiation Complex Assembly within the Wild-Strain of Coxsackievirus B3 and Live-Attenuated Sabin3-like IRESes during the Initiation of Translation. International Journal of Molecular Sciences. 2013; 14(3):4400-4418. https://doi.org/10.3390/ijms14034400
Chicago/Turabian StyleSouii, Amira, Manel Ben M'hadheb-Gharbi, Bruno Sargueil, Audrey Brossard, Nathalie Chamond, Mahjoub Aouni, and Jawhar Gharbi. 2013. "Ribosomal Initiation Complex Assembly within the Wild-Strain of Coxsackievirus B3 and Live-Attenuated Sabin3-like IRESes during the Initiation of Translation" International Journal of Molecular Sciences 14, no. 3: 4400-4418. https://doi.org/10.3390/ijms14034400



